Ehrenberg lab
Måns Ehrenberg’s research group focuses mainly on the prokaryotic ribosome and bacterial protein synthesis. The aim is to understand the mechanisms of initiation of protein synthesis, elongation of proteins and accuracy in tRNA and release factor selection, termination of protein synthesis and recycling of ribosomes from termination back to initiation – and the effects of antibiotics on all these mechanisms, as well as the overall effect on the growth rate of the cells. We use quantitative biochemical methods in combination with mathematical modelling, molecular genetics and in vitro experiments on bacteria.
Popular science presentation
The ribosome and its helping factors constitute the efficient machinery by which the cells construct all the proteins needed in life, both as simple unicellular organisms and as parts of complex multicellular organisms such as humans. The ribosomes are composed of two subunits, the small and the large subunit. The ribosomes link amino acids into long peptide chains, and the amino acid sequences determine the structures that the chains fold into, which in turn determine the function of the proteins in the cell.
The sequences of the peptide chains are coded in the genes in the DNA. In the process called transcription, RNA polymerase transcribe the information from the genes to messenger RNA (mRNA), which is positioned between the two subunits of the ribosome at the initiation of protein synthesis and read by the ribosome in order to construct a correct peptide chain.
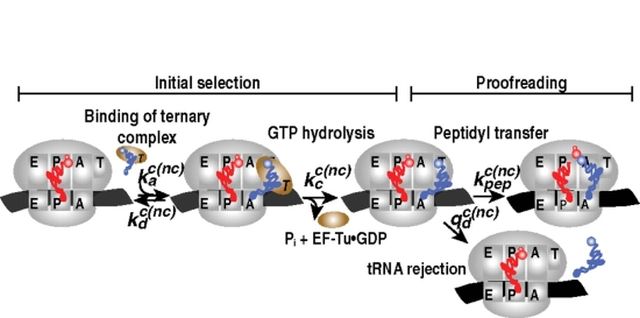
Each amino acid is chemically associated with one or more transport RNAs (tRNAs). They arrive at the ribosome in complex with the helping protein EF-Tu and decode the mRNA so that the amino acids are inserted in the correct order in the peptide chain. When an amino acid is incorporated in the peptide chain, the mRNA and tRNA with the aid of the helping protein EF-G are moved, or translocated, backwards by one code word within the ribosome, so that a new tRNA with a new amino acid can enter the ribosome and be linked to the growing peptide chain. This continues until the peptide chain is finished, and released from the ribosome using two termination factors. The ribosome is then recycled by separation of the two subunits so that it may translate the next protein. Again, the factor EF-G assists, together with the ribosomal recycling factor (RRF).
In our research we study every step of this entire process, mainly for bacterial protein synthesis but also some translation factors from eukaryotes (the evolutionary group that includes humans). The transcription of mRNA from DNA is described by mathematical models, in order to understand how the RNA polymerase can synthesize mRNA with the very high precision that has been observed. The initiation of protein synthesis and the repeated elongation of the peptide chains are studied using methods of biochemistry. A great interest of our group is how accurately the tRNAs can decode the mRNA, how this accuracy is achieved and how it improves or worsens by mutations, antibiotics or the surrounding conditions.
We are also interested in how the other processes of protein synthesis, such as initiation, translocation, termination and recycling, can be so fast and accurate. Previously, these processes could only be studied by “freezing” the ribosomes of the desired conformation, but the frozen complexes do not correspond to the authentic, functional complexes in the kinetic progress of the ribosomal functions. One solution is cryo-electron microscopy, which we utilize in collaboration with Columbia University, NY, USA. In another collaboration, with the University of Hamburg, we use a method to detect the position of ribosome on the mRNA in order to study the effect of antibiotics on translocation and recycling. We also use mathematical models and stochastic simulations to study the effect control systems of gene expression of growing bacteria, and how the control systems are affected by antibiotics.
Research projects
Protein synthesis
The goal is to understand the mechanisms of termination of protein synthesis, recycling of ribosomes from termination back to initiation, initiation of protein synthesis, elongation of proteins, accuracy of tRNA and release factor selection by the messengerRNA coded ribosome, toxicity of mini-genes and drop-off of peptidyl-tRNA. We use quantitative biochemical methods in combination with molecular genetics and experiments on living bacteria. Future directions include studying mechanisms for protein export and structural analysis of important, functional ribosome-factor complexes.
Protein synthesis in biotechnology
The goal is to constitute an in vitro system for bacterial protein synthesis which can be used to (i) produce any conceivable protein in large scale starting from its gene sequence (ii) synthesise proteins which are isotope labelled in chosen regions to facilitate structural analysis with NMR (iii) develop new, powerful techniques for combinatorial design of oligo-peptides and proteins and apply these methods to obtain new antibiotics, new protein biosensors and new catalysts.
Systems Biology
The Ehrenberg lab is actively exploring a number of topics in the area of Systems Biology.
- Stochastic modelling of copy number control for plasmids, with special reference to plasmids ColE1 and R1. The focus has been on how plasmids can achieve precise control of copy numbers to minimize plasmid losses at cell division also when their average copy numbers per cell are small.
- General features of noise in intracellular control systems. This area includes hyper-fluctuations in intracellular chemical reactions which operate near criticality and a suggestion, stochastic focusing, for how noise can enhance, rather than reduce sensitivity in molecular control systems.
- Regulation of protein synthesis, adaptation and growth control in bacteria. This area deals with how bacteria with the help of local control systems for regulation of gene expression (e.g. repressors and attenuation of transcription) in combination with global control systems (e.g. the stringent response to amino acid starvation) can grow fast in different media and adapt rapidly to environmental changes. Theoretical modelling and experimental approaches are combined.
- Action of antibiotics and mechanisms for antibiotic resistance in bacteria (in collaboration with Tanel Tenson, Tartu). This work is primarily dealing with the action of macrolides on bacterial protein synthesis and descriptions of a number of resistance mechanisms against these drugs. Theoretical modelling and experimental approaches are combined.
- Development of numerical methods for stochastic descriptions of intracellular reaction-networks including diffusion-reaction couplings (in collaboration with Per Lötstedt, Scientific Computing, Uppsala University). This work is dealing with developments of efficient algorithms for numerical solutions of the master equation in intracellular chemical networks. It also concerns descriptions of diffusion-reaction couplings in macroscopically bistable systems.
In collaboration with
- Tanel Tenson, Tartu University, Estonia. Group leader
- Per Lötstedt, Uppsala University, Sweden. Group leader
- Otto Berg, Uppsala University, Sweden. Emeritus
- Hans Bremer, University of Dallas, Texas, USA. Emeritus
- Zoya Ignatova, University of Hamburg, Germany. Group leader
Group members
Publications
Dynamics of release factor recycling during translation termination in bacteria
Part of Nucleic Acids Research, p. 5774-5790, 2023
- DOI for Dynamics of release factor recycling during translation termination in bacteria
- Download full text (pdf) of Dynamics of release factor recycling during translation termination in bacteria
Uncovering translation roadblocks during the development of a synthetic tRNA
Part of Nucleic Acids Research, p. 10201-10211, 2022
- DOI for Uncovering translation roadblocks during the development of a synthetic tRNA
- Download full text (pdf) of Uncovering translation roadblocks during the development of a synthetic tRNA
Estimation of peptide elongation times from ribosome profiling spectra
Part of Nucleic Acids Research, p. 5124-5142, 2021
- DOI for Estimation of peptide elongation times from ribosome profiling spectra
- Download full text (pdf) of Estimation of peptide elongation times from ribosome profiling spectra
Part of Nucleic Acids Research, p. 2684-2699, 2021
- DOI for N-6-Methyladenosines in mRNAs reduce the accuracy of codon reading by transfer RNAs and peptide release factors
- Download full text (pdf) of N-6-Methyladenosines in mRNAs reduce the accuracy of codon reading by transfer RNAs and peptide release factors
Part of mBio, 2019
- DOI for Reiterative Synthesis by the Ribosome and Recognition of the N-Terminal Formyl Group by Biosynthetic Machinery Contribute to Evolutionary Conservation of the Length of Antibiotic Microcin C Peptide Precursor
- Download full text (pdf) of Reiterative Synthesis by the Ribosome and Recognition of the N-Terminal Formyl Group by Biosynthetic Machinery Contribute to Evolutionary Conservation of the Length of Antibiotic Microcin C Peptide Precursor
Part of eLIFE, 2019
- DOI for The mechanism of error induction by the antibiotic viomycin provides insight into the fidelity mechanism of translation
- Download full text (pdf) of The mechanism of error induction by the antibiotic viomycin provides insight into the fidelity mechanism of translation
Part of Nature Communications, 2019
- DOI for The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy
- Download full text (pdf) of The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy
2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation
Part of Nature Structural & Molecular Biology, p. 208-216, 2018
Accuracy of genetic code translation and its orthogonal corruption by aminoglycosides and Mg2+ ions
Part of Nucleic Acids Research, p. 1362-1374, 2018
- DOI for Accuracy of genetic code translation and its orthogonal corruption by aminoglycosides and Mg2+ ions
- Download full text (pdf) of Accuracy of genetic code translation and its orthogonal corruption by aminoglycosides and Mg2+ ions
Part of Nucleic Acids Research, p. 5861-5874, 2018
- DOI for Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-Tu(H84A)
- Download full text (pdf) of Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-Tu(H84A)
How 2 '-O-Methylation in mRNA Disrupts tRNA Decoding during Translation Elongation
Part of Biophysical Journal, 2018
The Structural Basis for Initiation Factor 2 Activation during Translation Initiation
Part of Biophysical Journal, 2018
Part of Nature Communications, 2017
- DOI for A conformational switch in initiation factor 2 controls the fidelity of translation initiation in bacteria
- Download full text (pdf) of A conformational switch in initiation factor 2 controls the fidelity of translation initiation in bacteria
Ribosomes are optimized for autocatalytic production
Part of Nature, p. 293-297, 2017
Transcriptional accuracy modeling suggests two-step proofreading by RNA polymerase
Part of Nucleic Acids Research, p. 11582-11593, 2017
- DOI for Transcriptional accuracy modeling suggests two-step proofreading by RNA polymerase
- Download full text (pdf) of Transcriptional accuracy modeling suggests two-step proofreading by RNA polymerase
Complete kinetic mechanism for recycling of the bacterial ribosome
Part of RNA, p. 10-21, 2016
Key Intermediates in Ribosome Recycling Visualized by Time-Resolved Cryoelectron Microscopy
Part of Structure, p. 2092-2101, 2016
Part of Nucleic Acids Research, p. 3264-3275, 2016
- DOI for Mechanism of fusidic acid inhibition of RRF- and EF-G-dependent splitting of the bacterial post-termination ribosome
- Download full text (pdf) of Mechanism of fusidic acid inhibition of RRF- and EF-G-dependent splitting of the bacterial post-termination ribosome
Molecular mechanism of viomycin inhibition of peptide elongation in bacteria
Part of Proceedings of the National Academy of Sciences of the United States of America, p. 978-983, 2016
N-6-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics
Part of Nature Structural & Molecular Biology, p. 110-+, 2016
Proofreading neutralizes potential error hotspots in genetic code translation by transfer RNAs
Part of RNA, p. 896-904, 2016
- DOI for Proofreading neutralizes potential error hotspots in genetic code translation by transfer RNAs
- Download full text (pdf) of Proofreading neutralizes potential error hotspots in genetic code translation by transfer RNAs
Two proofreading steps amplify the accuracy of genetic code translation
Part of Proceedings of the National Academy of Sciences of the United States of America, p. 13744-13749, 2016
Accuracy of initial codon selection by aminoacyl-tRNAs on the mRNA-programmed bacterial ribosome
Part of Proceedings of the National Academy of Sciences of the United States of America, p. 9602-9607, 2015
- DOI for Accuracy of initial codon selection by aminoacyl-tRNAs on the mRNA-programmed bacterial ribosome
- Download full text (pdf) of Accuracy of initial codon selection by aminoacyl-tRNAs on the mRNA-programmed bacterial ribosome
Determinants of the Rate of mRNA Translocation in Bacterial Protein Synthesis
Part of Journal of Molecular Biology, p. 1835-1847, 2015
- DOI for Determinants of the Rate of mRNA Translocation in Bacterial Protein Synthesis
- Download full text (pdf) of Determinants of the Rate of mRNA Translocation in Bacterial Protein Synthesis
DNA Template Dependent Accuracy Variation of Nucleotide Selection in Transcription
Part of PLOS ONE, 2015
- DOI for DNA Template Dependent Accuracy Variation of Nucleotide Selection in Transcription
- Download full text (pdf) of DNA Template Dependent Accuracy Variation of Nucleotide Selection in Transcription
Free RNA polymerase in Escherichia coli
Part of Biochimie, p. 80-91, 2015
Part of Journal of Biological Chemistry, p. 3440-3454, 2015
On the pH Dependence of Class-1 RF-Dependent Termination of mRNA Translation
Part of Journal of Molecular Biology, p. 1848-1860, 2015
- DOI for On the pH Dependence of Class-1 RF-Dependent Termination of mRNA Translation
- Download full text (pdf) of On the pH Dependence of Class-1 RF-Dependent Termination of mRNA Translation
A tRNA body with high affinity for EF-Tu hastens ribosomal incorporation of unnatural amino acids
Part of RNA, p. 632-643, 2014
- DOI for A tRNA body with high affinity for EF-Tu hastens ribosomal incorporation of unnatural amino acids
- Download full text (pdf) of A tRNA body with high affinity for EF-Tu hastens ribosomal incorporation of unnatural amino acids
Peptide Formation by N-Methyl Amino Acids in Translation Is Hastened by Higher pH and tRNAPro
Part of ACS Chemical Biology, p. 1303-1311, 2014
Thermodynamic Modeling of Variations in the Rate of RNA Chain Elongation of E-coli rrn Operons
Part of Biophysical Journal, p. 55-64, 2014
Cryo-EM visualization of the ribosome in termination complex with apo-RF3 and RF1
Part of eLife, 2013
- DOI for Cryo-EM visualization of the ribosome in termination complex with apo-RF3 and RF1
- Download full text (pdf) of Cryo-EM visualization of the ribosome in termination complex with apo-RF3 and RF1
Optimal control of gene expression for fast proteome adaptation to environmental change
Part of Proceedings of the National Academy of Sciences of the United States of America, p. 20527-20532, 2013
Optimal Strategy for Rapid Proteome Re-Arrangements in Bacterial Populations
Part of Biophysical Journal, 2013
The Impact of Aminoglycosides on the Dynamics of Translation Elongation
Part of Cell Reports, p. 497-508, 2013
Part of Proceedings of the National Academy of Sciences of the United States of America, p. 131-136, 2012
Inefficient delivery but fast peptide bond formation of unnatural l -aminoacyl-tRNAs in translation
Part of Journal of the American Chemical Society, p. 17955-17962, 2012
Positive allosteric feedback regulation of the stringent response enzyme RelA by its product
Part of EMBO Reports, p. 835-839, 2012
Activation of initiation factor 2 by ligands and mutations for rapid docking of ribosomal subunits
Part of EMBO Journal, p. 289-301, 2011
Comment on "The mechanism for activation of GTP hydrolysis on the ribosome"
Part of Science, p. 37, 2011
Identification of enzyme inhibitory mechanisms from steady-state kinetics
Part of Biochimie, p. 1623-1629, 2011
Part of Proceedings of the National Academy of Sciences of the United States of America, p. 79-84, 2011
Error-prone initiation factor 2 mutations reduce the fitness cost of antibiotic resistance
Part of Molecular Microbiology, p. 1299-1313, 2010
- DOI for Error-prone initiation factor 2 mutations reduce the fitness cost of antibiotic resistance
- Download full text (pdf) of Error-prone initiation factor 2 mutations reduce the fitness cost of antibiotic resistance
Part of Biochimie, p. 12-20, 2010
Protein synthesis: Translocation in slow motion
Part of Nature, p. 325-326, 2010
Ribosomes lacking protein S20 are defective in mRNA binding and subunit association
Part of Journal of Molecular Biology, p. 767-776, 2010
Part of Journal of Molecular Biology, p. 838-846, 2010
tmRNA-SmpB complex mimics native aminoacyl-tRNAs in the A site of stalled ribosomes.
Part of Journal of Structural Biology, p. 342-348, 2010
Cis-acting resistance peptides reveal dual ribosome inhibitory action of the macrolide josamycin
Part of Biochimie, p. 989-995, 2009
Drug efflux pump deficiency and drug target resistance masking in growing bacteria
Part of Proceedings of the National Academy of Sciences of the United States of America, p. 8215-8220, 2009

