Sanyal lab
Prof. Suparna Sanyal’s laboratory studies mechanisms of protein synthesis and protein folding using mainly molecular biology, biochemistry and structural biology tools. We frequently use fast kinetics such as quench-flow and stopped-flow for solving mechanistic problems. These are complemented with structures solved with X-ray crystallography and cryo-EM.Our current research topics include mechanisms of different steps of translation in bacteria and eukaryotes, function of the ribosomal proteins - particularly the ‘ribosomal stalk’, antibiotics and antibiotics resistance, translation in pathogenic microbes (Mycobacteria, Staphylococcus, Plasmodium, Giardia etc.), translational GTPases, ribosome biogenesis and evolution of translation machinery.We have developed a reconstituted transcription-translation-folding system (RTTF) that produces active proteins in the test tube. We are pioneers to show protein folding activity of the ribosome (PFAR) and recently discovered a link between PFAR and prion diseases.
Popular science presentation
Protein Synthesis & Protein Folding : Mechanisms, Diseases and Biotechnology
The Ribosome is the protein factory of the cell, which translates genetic codes from messenger RNA (mRNA) to proteins. Protein synthesis also requires many translation factors, which play important roles in the process. In our lab, we are trying to understand how the ribosome and the translation factors work, how they carry out the different stages of protein synthesis. Although we perform most of the experiments in the test tube using advanced techniques of biochemistry and molecular biology, these give us some idea about the protein synthesis process in the cell. Moreover, these allow us to understand how antibiotics, which we often get prescribed from doctors for treating bacterial infections, work to block protein synthesis in bacterial cells. These studies are particularly important for fighting drug-resistant bacteria causing severe diseases such as tuberculosis. In the future, these studies may also aid in designing new drugs against pathogenic microbes. In addition, using individual components of the bacterial protein synthesis machinery, we can now produce various peptides and proteins in the test tube, which is the first step towards constructing an artificial cell.
The peptide chains need to fold properly to gain their function. We have demonstrated that ribosomes can assist in protein folding by using its RNA of the large subunit. Recently, we have found that the protein folding activity of the ribosome can be linked to prion diseases, which include scrapie, mad cow disease, Creutzfeldt–Jacob disease etc. Prion proteins are infectious proteins, which when misfolded can form amyloid fibrils and can damage brain function. We are now studying the role of the ribosomal RNA in folding of prion proteins, which will allow us to develop strategies to combat these fatal diseases.
Research projects
The role of the ribosomal stalk protein L7/L12 in the interaction with translation factors
Chandra Sekhara Mandava, Ph.D.
Xueliang Ge, Ph.D.
Ranjeet Kumar, Chenhui Huang, Josefine Ederth, Former Postdocs
Christiane Lupczyk, Former Project Student
In bacteria, the ribosomal 'stalk' consists of two L7/L12 protein dimers and one L10 protein attached to the rRNA next to the protein L11. The L7/L12 proteins are highly flexible and change conformation in different steps of translation. These proteins interact with the translational G-factors such as EF-G and EF-Tu, and may be involved in GTP binding and in activation of factor dependent GTP hydrolysis. However, the molecular mechanism of these processes is quite unclear. The specific goal of this project is to characterize the role of L7/L12 in interactions with translational G-proteins. Using heteronuclear NMR, we have mapped the residues on L7/L12 that are involved directly in interaction with IF2, EF-G, EF-Tu and RF3. We will also map the regions on the factors involved in these interactions as the next step. Another aim is to study whether L7/L12 acts as GTPase activator for IF2 and RF3 as it does for EF-G and EF-Tu. We are also studying the effect of depletion of ribosomal stalk proteins (L7/L12, L8 and L11) in IF2 dependent subunit association.We have produced an E. coli strain, which has ribosomes with only one L7/L12 dimer. In vivo and in vitro characterization of these ribosomes are ongoing.
The fusidic acid resistance in Staphylococcus aureus.
Ravi Kiran Koripella, Ph.D. Student
Mikael Holm, Ph.D Student
Hasanthi Karunasekera, Jenny Honek Former Project Students
Chenhui Huang, Musturi Venkataramana, Former Postdocs
Key investigator: Suparna Sanyal
Fusidic acid is an antibiotic, clinically used to prevent Staphylococcus aureus infection since last four decades. It binds to EF-G and blocks EF-G.GDP on the ribosome in the post-translocation state. So far all fusidic acid related studies are done with E. coli and some other thermophilic bacteria, which are not the natural targets for fusidic acid and it is practically impermeable in these hosts. This is the first concerted effort to study the fusidic acid inhibition and resistance mechanism in vitro using the EF-Gs from Staphylococcus aureus, the actual drug target of fusidic acid. Our goals are a)To locate the fusidic acid binding site on EF-G. b)To characterize the fusidic acid resistant mutants of Staphylococcus aureus, particularly those which have gained biological fitness by additional mutations in the fusA gene.
Ribosome assisted protein folding and it's implication in prion diseases
Yanhong Pang, Ph.D. Student
Petar Kovachev, Ph.D. Student
Debapriya Banerjee, Ph.D.
Venkata Sriram Kurella Ariane Schabe, Neelanjan Vishnu Former Project students
Suzana DosReis, Former Postdoc
Debasis Das, Erik Richter, Helena Kristiansson, Hiroki Kawahara Former Project /Visiting Students
Collaboration with Marc Blondel Group, Brest, France
Recently our understanding of interplay between structure and function of bacterial ribosome has increased significantly. One major question that has remained unanswered, is how newly synthesized polypeptide chains are folded in the living cells. Several molecular chaperones have been shown to be part of the process. But it has also been demonstrated in vitro that ribosomes from archaebacteria, eubacteria, animals, plants and mitochondria can refold a large number of denatured proteins to their active state all the different proteins which have been subjected to ribosomal refolding show remarkable activity recovery compared to spontaneous folding (Das B et al, 1996). The ribosome assisted protein folding activity have been assigned to the large subunit 50S, and more precisely to the domain V of the 23SrRNA. The aim of the project is to try to understand how this folding occurs, is it co or posttranslational? Does this folding happen on the translating ribosome or in trans? Is there any universal binding motif on peptides that are recognized by the ribosome? Is there any implication of this function in prion-diseases?
The prions are infectious proteins that cause a group of fatal neurodegenerative disease like Creutzfeld-Jacob or BSE (Mad Cow Disease). These diseases involve the conversion of the normal form of the protein (PrPc) into the disease causing isoform (PrPsc) and are recognized as a 'protein-folding associated problem'. Recently we have shown in collaboration with Marc Blondel group in France, that two antiprion drugs 6AP and GA use ribosomes as the molecular target in the cell (Tribouillard- Tanvier D et al, 2008). These drugs bind specifically to ribosomal RNA from the large subunit and inhibit the folding activity of the ribosome, but not protein synthesis. The detailed characterization of the process is ongoing.
In a recent in vitro study it has been shown that the unfolded proteins can dissociate 70S ribosome into the subunits while refolding. In another study, the nascent protein chain is seen to be associated with the 50S subunit immediately after its synthesis. This implicates that the nascent protein chain may have a role in translation termination and the dissociation of the ribosomal subunits. At present we are studying the release and folding of a full-length protein chain using a cell-free protein synthesis system.
Translation termination in bacteria
Xueliang Ge, Ph.D
Gürkan Korkmaz, Ph.D Student
Ravi Kiran Koripella, Ph.D
Chenhui Huang, Former Postdoc
Nhan Tran Van, Anders Bankefors, Chen Chen Former Project Student
Collaboration with Joachim Frank Group, New York, USA and Valerie Heurgue-Hamard, CNRS Paris, France
In the termination process of translation the newly synthesized protein is being released from the ribosome. In this step of translation two class I release factor (RF) overlap partly in their codon recognition and are being recycled by a class II release factor namely RF3.
We are approaching the mechanism of translation termination by investigating the action of RF3 involving possible interaction with RF1/2 and ribosomal protein L12. Further we are interested in the decoding of stop codons by class I release factors. Recently a computational approach was used to simulate and calculate the binding energy of several amino acids of RF1/2, in this project we want to compare these results with our experimental mutagenesis. Additionally we published a database analyzing the stop codon usage in bacteria.
Translation in Mycobacteria
Xueliang Ge Ph.D
Emelie Gabrielsson Former Master Student
Ranjeet Kumar Former Postdoc
Translocation and RNA
Mikael Holm, Ph.D Student
Reconstituted Transcription Translation Folding System
Kristin Peisker, Ph.D
Sunanda Chatterjee, PhD
Jan Pille, Project Student, Antje Wanglin former M. Sc. Research Assistant
Reconstituted Transcription Translation folding (RTTF) system for cell free protein synthesis:
We have been successful to reconstitute a cell-free transcription-translation-folding system (named as RTTF system) using purified translation components from bacteria E. coli, where active proteins are being synthesized with good efficiency. So far, we have produced firefly-luciferase, dihydrofolate reductase (DHFR), fluorescent proteins mCherry and turbo-GFP in the system starting from DNA or in vitro transcribed mRNA. The system provides great flexibility to test individual translational components with mutations or with specific modulators. Some examples are ribosomes with altered stalk protein complex (Mandava et al., NAR, 2012); L1 deleted ribosome (Tobin et al., under preparation) and fusidic acid resistant mutant EF-Gs from pathogenic bacteria Staphylococcus aureus. Further, the system can be used for checking accuracy in protein synthesis, frame-shifting defects as well as for studying refolding and maturation of the target proteins. Currently, a similar system with Mycobacterial translation machinery is under construction, which will be used for high-throughput screening of potential TB drugs. Last but not the least, this system provides ample scope to produce unique peptides and engineered proteins which can be of great value for biomedical and technical applications.
The system has been highly evaluated by the international expert panel appointed by The Swedish Research Council and Formas. Read the comment quoted from the Draft Midterm Evaluation Report of the 2006 Linnaeus environments and Doctoral programs for UPPSALA RNA RESEARCH CENTER
For exam-work on related topics please contact suparna.sanyal@icm.uu.se
Group members
Publications
Part of Scientific Reports, 2024
- DOI for Structures of the Staphylococcus aureus ribosome inhibited by fusidic acid and fusidic acid cyclopentane
- Download full text (pdf) of Structures of the Staphylococcus aureus ribosome inhibited by fusidic acid and fusidic acid cyclopentane
Part of Nature Communications, 2023
- DOI for Antibiotic thermorubin tethers ribosomal subunits and impedes A-site interactions to perturb protein synthesis in bacteria
- Download full text (pdf) of Antibiotic thermorubin tethers ribosomal subunits and impedes A-site interactions to perturb protein synthesis in bacteria
Part of Nucleic Acids Research, p. 3436-3451, 2023
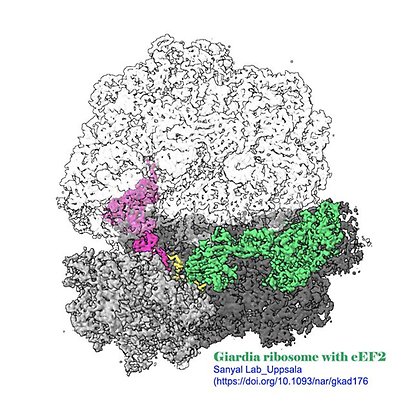
- DOI for Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis
- Download full text (pdf) of Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis
Part of RNA Biology, p. 681-692, 2023
- DOI for Lamotrigine compromises the fidelity of initiator tRNA recruitment to the ribosomal P-site by IF2 and the RbfA release from 30S ribosomes in Escherichia coli
- Download full text (pdf) of Lamotrigine compromises the fidelity of initiator tRNA recruitment to the ribosomal P-site by IF2 and the RbfA release from 30S ribosomes in Escherichia coli
Molecular basis of the pleiotropic effects by the antibiotic amikacin on the ribosome
Part of Nature Communications, 2023
- DOI for Molecular basis of the pleiotropic effects by the antibiotic amikacin on the ribosome
- Download full text (pdf) of Molecular basis of the pleiotropic effects by the antibiotic amikacin on the ribosome
Part of iScience, 2021
- DOI for Cotranslational recruitment of ribosomes in protocells recreates a translocon-independent mechanism of proteorhodopsin biogenesis
- Download full text (pdf) of Cotranslational recruitment of ribosomes in protocells recreates a translocon-independent mechanism of proteorhodopsin biogenesis
Part of Journal of Biological Chemistry, 2021
- DOI for GGQ methylation enhances both speed and accuracy of stop codon recognition by bacterial class-I release factors
- Download full text (pdf) of GGQ methylation enhances both speed and accuracy of stop codon recognition by bacterial class-I release factors
Part of Molecular biology and evolution, p. 3436-3444, 2021
Mechanistic insights into translation inhibition by aminoglycoside antibiotic arbekacin
Part of Nucleic Acids Research, p. 6880-6892, 2021
Part of RNA Biology, p. 2363-2375, 2021
- DOI for Optimization of a fluorescent-mRNA based real-time assay for precise kinetic measurements of ribosomal translocation
- Download full text (pdf) of Optimization of a fluorescent-mRNA based real-time assay for precise kinetic measurements of ribosomal translocation
Real-time measurements of aminoglycoside effects on protein synthesis in live cells
Part of Proceedings of the National Academy of Sciences of the United States of America, 2021
- DOI for Real-time measurements of aminoglycoside effects on protein synthesis in live cells
- Download full text (pdf) of Real-time measurements of aminoglycoside effects on protein synthesis in live cells
Repurposing tRNAs for nonsense suppression
Part of Nature Communications, 2021
- DOI for Repurposing tRNAs for nonsense suppression
- Download full text (pdf) of Repurposing tRNAs for nonsense suppression
Collateral toxicity limits the evolution of bacterial Release Factor 2 towards total omnipotence
Part of Molecular biology and evolution, p. 2918-2930, 2020
- DOI for Collateral toxicity limits the evolution of bacterial Release Factor 2 towards total omnipotence
- Download full text (pdf) of Collateral toxicity limits the evolution of bacterial Release Factor 2 towards total omnipotence
Part of Proceedings of the National Academy of Sciences of the United States of America, p. 15609-15619, 2020
Ribosomal RNA Modulates Aggregation of thePodosporaPrion Protein HET-s
Part of International Journal of Molecular Sciences, 2020
- DOI for Ribosomal RNA Modulates Aggregation of thePodosporaPrion Protein HET-s
- Download full text (pdf) of Ribosomal RNA Modulates Aggregation of thePodosporaPrion Protein HET-s
Selective translation by alternative bacterial ribosomes
Part of Proceedings of the National Academy of Sciences of the United States of America, p. 19487-19496, 2020
- DOI for Selective translation by alternative bacterial ribosomes
- Download full text (pdf) of Selective translation by alternative bacterial ribosomes
Inhibition of translation termination by small molecules targeting ribosomal release factors
Part of Scientific Reports, 2019
- DOI for Inhibition of translation termination by small molecules targeting ribosomal release factors
- Download full text (pdf) of Inhibition of translation termination by small molecules targeting ribosomal release factors
RNA modulates aggregation of the recombinant mammalian prion protein by direct interaction
Part of Scientific Reports, 2019
- DOI for RNA modulates aggregation of the recombinant mammalian prion protein by direct interaction
- Download full text (pdf) of RNA modulates aggregation of the recombinant mammalian prion protein by direct interaction
Part of eLIFE, 2019
- DOI for The mechanism of error induction by the antibiotic viomycin provides insight into the fidelity mechanism of translation
- Download full text (pdf) of The mechanism of error induction by the antibiotic viomycin provides insight into the fidelity mechanism of translation
Part of The Journal of Physical Chemistry C, p. 21031-21038, 2018
Part of Advanced Synthesis and Catalysis, p. 3930-3939, 2018
Part of Proceedings of the National Academy of Sciences of the United States of America, p. 4649-4654, 2018
Part of Nucleic Acids Research, p. 5861-5874, 2018
- DOI for Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-Tu(H84A)
- Download full text (pdf) of Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-Tu(H84A)
Part of Protein & Cell, p. 384-388, 2018
- DOI for Cryo-EM structure of Mycobacterium smegmatis ribosome reveals two unidentified ribosomal proteins close to the functional centers
- Download full text (pdf) of Cryo-EM structure of Mycobacterium smegmatis ribosome reveals two unidentified ribosomal proteins close to the functional centers
A genetically encoded fluorescent tRNA is active in live-cell protein synthesis
Part of Nucleic Acids Research, p. 4081-4093, 2017
- DOI for A genetically encoded fluorescent tRNA is active in live-cell protein synthesis
- Download full text (pdf) of A genetically encoded fluorescent tRNA is active in live-cell protein synthesis
Distinct modulatory role of RNA in the aggregation of the tumor suppressor protein p53 core domain.
Part of Journal of Biological Chemistry, p. 9345-9357, 2017
Experimental Evolution of Escherichia coli Harboring an Ancient Translation Protein
Part of Journal of Molecular Evolution, p. 69-84, 2017
- DOI for Experimental Evolution of Escherichia coli Harboring an Ancient Translation Protein
- Download full text (pdf) of Experimental Evolution of Escherichia coli Harboring an Ancient Translation Protein
Part of npj Biofilms and Microbiomes, 2017
- DOI for New melanocortin-like peptide of E. coli can suppress inflammation via the mammalian melanocortin-1 receptor (MC1R): possible endocrine-like function for microbes of the gut.
- Download full text (pdf) of New melanocortin-like peptide of E. coli can suppress inflammation via the mammalian melanocortin-1 receptor (MC1R): possible endocrine-like function for microbes of the gut.
Part of Journal of Biological Chemistry, p. 15134-15142, 2017
Regulation of PfEMP1-VAR2CSA translation by a Plasmodium translation-enhancing factor
Part of Nature Microbiology, 2017
Spatial Distribution and Ribosome-Binding Dynamics of EF-P in Live Escherichia coli
Part of mBio, 2017
- DOI for Spatial Distribution and Ribosome-Binding Dynamics of EF-P in Live Escherichia coli
- Download full text (pdf) of Spatial Distribution and Ribosome-Binding Dynamics of EF-P in Live Escherichia coli
Micrococcin P1-A bactericidal thiopeptide active against Mycobacterium tuberculosis
Part of Tuberculosis, p. 95-101, 2016
Molecular mechanism of viomycin inhibition of peptide elongation in bacteria
Part of Proceedings of the National Academy of Sciences of the United States of America, p. 978-983, 2016
A conserved histidine in switch-II of EF-G moderates release of inorganic phosphate
Part of Scientific Reports, 2015
- DOI for A conserved histidine in switch-II of EF-G moderates release of inorganic phosphate
- Download full text (pdf) of A conserved histidine in switch-II of EF-G moderates release of inorganic phosphate
Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G
Part of Science Advances, 2015
- DOI for Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G
- Download full text (pdf) of Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G
Part of International Journal of Quantum Chemistry, p. 846-852, 2015
Part of Journal of Biological Chemistry, p. 3440-3454, 2015
HflX is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions
Part of Nature Structural & Molecular Biology, p. 906-913, 2015
Characterizing an engineered release factor capable of reading all three stop codons
Part of The FASEB Journal, 2014
Part of Journal of Biological Chemistry, p. 30334-30342, 2014
Organization of Ribosomes and Nucleoids in Escherichia coli Cells during Growth and in Quiescence
Part of Journal of Biological Chemistry, p. 11342-11352, 2014
Part of Biochimie, p. 194-199, 2014
Structural and Functional Insights into the Mode of Action of a Universally Conserved Obg GTPase
Part of PLoS biology, 2014
- DOI for Structural and Functional Insights into the Mode of Action of a Universally Conserved Obg GTPase
- Download full text (pdf) of Structural and Functional Insights into the Mode of Action of a Universally Conserved Obg GTPase
Part of ACS Chemical Neuroscience, p. 1075-1082, 2014
Cryo-EM visualization of the ribosome in termination complex with apo-RF3 and RF1
Part of eLife, 2013
- DOI for Cryo-EM visualization of the ribosome in termination complex with apo-RF3 and RF1
- Download full text (pdf) of Cryo-EM visualization of the ribosome in termination complex with apo-RF3 and RF1
Interaction of Nucleobases and Aromatic Amino Acids with Graphene Oxide and Graphene Flakes
Part of The Journal of Physical Chemistry Letters, p. 3710-3718, 2013
Part of Journal of Biological Chemistry, p. 19081-19089, 2013
The Toll-Like Receptor Agonist Imiquimod Is Active against Prions
Part of PLOS ONE, 2013
- DOI for The Toll-Like Receptor Agonist Imiquimod Is Active against Prions
- Download full text (pdf) of The Toll-Like Receptor Agonist Imiquimod Is Active against Prions
Part of Nucleic Acids Research, p. 2054-2064, 2012
- DOI for Bacterial ribosome requires multiple L12 dimers for efficient initiation and elongation of protein synthesis involving IF2 and EF-G
- Download full text (pdf) of Bacterial ribosome requires multiple L12 dimers for efficient initiation and elongation of protein synthesis involving IF2 and EF-G
Mechanism and Rates of Exchange of L7/L12 between Ribosomes and the Effects of Binding EF-G
Part of ACS Chemical Biology, p. 1120-1127, 2012
People
Suparna Sanyal, PhD, Docent
Professor and Head of the research program - Molecular Biology; Vice prefect - Department of Cell and Molecular Biology; PhD Education Responsible Professor (FUAP) - Molecular Life Sciences;
Director of the Master Program - Applied Biotechnology
Chandra Sekhar Mandava, PhD, Senior Researcher
affiliated to Testa Center as Uppsala University representative
Xueliang Ge, PhD Senior Researcher
Soneya Majumdar, PhD Postdoctoral Researcher
Liuqun Zhao, PhD, Postdoctoral Researcher
Narayan Parajuli, PhD student
Shreya Pundir, PhD student
Andrew Emmerich, PhD student
Arindam Tarafder, PhD student
Wei Guo, Master thesis student, Cell & Molecular Biology
Raymond Fowler, Research Technician and Molecular Biology lab Manager

