Griese lab
We use a combination of molecular biology, biochemical and structural biology methods, including X-ray crystallography and single-particle cryo-EM, to investigate metal homeostasis regulation in a bacterial model organism, the actinomycete Saccharopolyspora erythraea. We aim to obtain a complete understanding from the regulatory networks to the underlying molecular mechanisms.
Popular science presentation
Life is not possible without metals. Metal ions stabilize the structure of proteins and nucleic acids and catalyze chemical reactions that enzymes alone cannot accomplish. About half of all enzymes contain metal cofactors. Nature uses mainly the transition metals manganese, iron, cobalt, nickel, copper and zinc. These metals have different properties and fulfil different functions in the cell. Any imbalance in the cellular metal contents can lead to metals binding to the wrong enzymes, rendering these enzymes non-functional. Therefore, cells carefully balance metal uptake, storage and export so that their demands for each particular metal are exactly met. This process is called metal homeostasis.
In bacteria, intracellular metal concentrations are regulated by specialized transcription factors. These proteins recognize and bind to specific metals and specific DNA sequences, thereby either blocking or enhancing the expression of neighbouring genes. Thus, when manganese is bound to a sensor responsible for regulating manganese uptake, the sensor blocks transcription of genes encoding for manganese importers, so that no more importers are made when free manganese is already present in the cell.
We study the metal sensors from the bacterium Saccharopolyspora erythraea using different biochemical, biophysical and molecular biology methods. Presently we are investigating how they recognize their DNA target sites, aiming to understand the underlying molecular mechanism. Ultimately we want to unravel the complex regulatory networks these metal sensors control.
S. erythraea is best known for being the producer of erythromycin, an important antibiotic. S. erythraea and related species from the actinomycete family produce a large variety of compounds such as erythromycin that are called secondary metabolites and are often potent antibiotics. However, most strains do not produce much or any of the metabolites they can make under laboratory conditions. Many interesting compounds therefore remain to be discovered. Since the cellular metal status has a profound influence on metabolism, our research may help the continuing efforts to improve secondary metabolite production in S. erythraea and other actinomycetes.
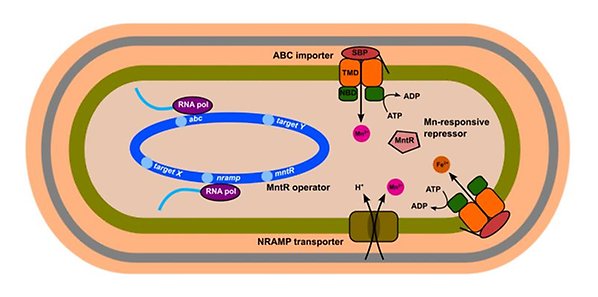
Illustration of metal homeostasis in gram-positive bacteria such as S. erythraea, using the example of manganese. Different types of membrane importers pump manganese into the cell. When the intracellular manganese concentration reaches a certain level, manganese binds to the manganese-responsive repressor MntR and activates it, so that it can bind to the promoters of genes involved in manganese metabolism, such as those encoding manganese importers, and repress their transcription.
Research projects
The overall goal of our research is to establish the regulatory networks controlling metal homeostasis in Saccharopolyspora erythraea, an actinomycete best known for being the producer of the macrolide antibiotic erythromycin. To obtain a comprehensive understanding, we aim to map out the regulatory networks as well as dissect the underlying molecular mechanisms.
Tight regulation of intracellular metal ion concentrations is crucial for any organism’s survival. The key players in prokaryotic metal homeostasis are metal-responsive transcriptional regulators. These metal sensors control the expression of genes encoding proteins responsible for metal uptake, efflux and storage, as well as metalloenzymes, to adjust metabolism in accordance with the cellular metal status.
We are currently investigating the metal sensors from S. erythraea, addressing two main questions:
1. How do the metal sensors recognize their target sites in DNA?
The fundamental mechanisms underlying protein–DNA recognition are incompletely understood. Although it is well-established that eukaryotic transcription factors recognize their target sites in the genome using a combination of interactions with specific DNA bases (base readout) and recognition of the DNA shape (shape readout), it remains unclear how transcription factors specifically recognize subtly different sequence-dependent DNA shapes. Moreover, the contribution of shape readout to target recognition has so far largely been overlooked when it comes to prokaryotic transcription factors. However, it is beginning to emerge that shape readout plays an important role even in these less complex systems (Marcos-Torres et al., NAR 2021). A central aim of our research is to fill in the gaps in our understanding of this fundamental molecular recognition mechanism. The relatively simple bacterial model systems that the metal sensors constitute allow us to dissect their protein–DNA recognition mechanism in detail and obtain a deeper molecular mechanistic understanding that will help us to understand the overarching principles of DNA shape readout.
2. Which genes do the metal sensors regulate?
Metal homeostasis has been intensively studied for actinomycete pathogens, in particular of the genera Mycobacterium and Corynebacterium, where the iron sensor IdeR/DtxR is required not only to control iron homeostasis, but also to maintain virulence. In contrast, metal homeostasis networks are not well described for antibiotic-producing actinomycetes such as S. erythraea. These networks can be expected to influence the production of medically interesting secondary metabolites such as erythromycin. Although much effort has been made to improve production of this secondary metabolite, the complex regulation of this process is still not completely understood. Our research will contribute to a better understanding of how primary and secondary metabolism are regulated in S. erythraea, ultimately enabling an improvement of secondary metabolite production and potentially the discovery of new medically interesting metabolites.
To address these questions, we use molecular and microbiology methods, recombinant protein production and purification, X-ray crystallography, small-angle X-ray scattering (SAXS), single-particle cryogenic electron microscopy (cryo-EM) and nuclear magnetic resonance spectroscopy (NMR), as well as different biochemical and biophysical techniques to study DNA binding, such as electrophoretic mobility shift assays (EMSAs) and fluorescence spectroscopy.
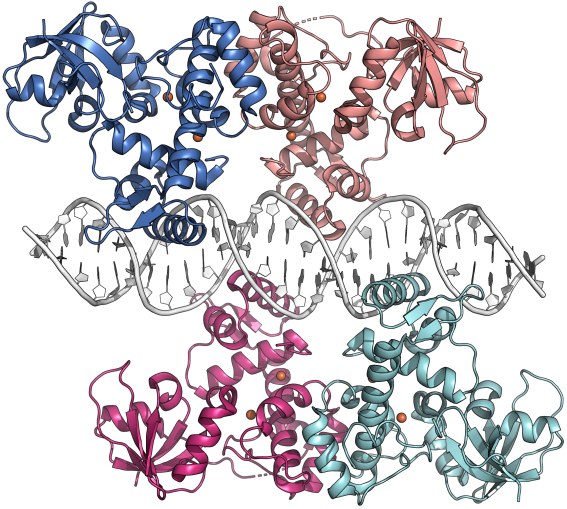
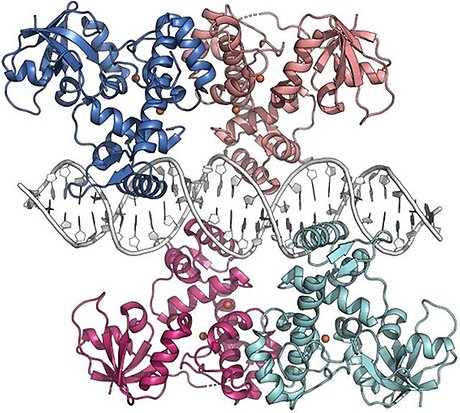
Crystal structure of the S. erythraea iron-dependent regulator IdeR in complex with iron and its consensus DNA recognition sequence (PDB ID 7B20; Marcos-Torres et al., NAR 2021). Each IdeR subunit binds two metal ions in adjacent binding sites. Two IdeR dimers bind to the palindromic DNA recognition sequence.
Group members
Publications
Computational design of the temperature optimum of an enzyme reaction
Part of Science Advances, 2023
- DOI for Computational design of the temperature optimum of an enzyme reaction
- Download full text (pdf) of Computational design of the temperature optimum of an enzyme reaction
Part of FEBS Letters, p. 1600-1610, 2022
- DOI for Comparative structural analysis provides new insights into the function of R2‐like ligand‐binding oxidase
- Download full text (pdf) of Comparative structural analysis provides new insights into the function of R2‐like ligand‐binding oxidase
The bacterial iron sensor IdeR recognizes its DNA targets by indirect readout
Part of Nucleic Acids Research, 2021
- DOI for The bacterial iron sensor IdeR recognizes its DNA targets by indirect readout
- Download full text 1 (pdf) of The bacterial iron sensor IdeR recognizes its DNA targets by indirect readout
- Download full text 2 (pdf) of The bacterial iron sensor IdeR recognizes its DNA targets by indirect readout
Part of Journal of the American Chemical Society, p. 5338-5354, 2020
- DOI for Key Structural Motifs Balance Metal Binding and Oxidative Reactivity in a Heterobimetallic Mn/Fe Protein
- Download full text (pdf) of Key Structural Motifs Balance Metal Binding and Oxidative Reactivity in a Heterobimetallic Mn/Fe Protein
Part of Journal of Biological Inorganic Chemistry, p. 571-582, 2020
- DOI for The Bacillus anthracis class Ib ribonucleotide reductase subunit NrdF intrinsically selects manganese over iron
- Download full text (pdf) of The Bacillus anthracis class Ib ribonucleotide reductase subunit NrdF intrinsically selects manganese over iron
Part of Journal of Biological Inorganic Chemistry, p. 211-221, 2019
- DOI for Assembly of a heterodinuclear Mn/Fe cofactor is coupled to tyrosine-valine ether cross-link formation in the R2-like ligand-binding oxidase
- Download full text (pdf) of Assembly of a heterodinuclear Mn/Fe cofactor is coupled to tyrosine-valine ether cross-link formation in the R2-like ligand-binding oxidase
Chemical flexibility of heterobimetallic Mn/Fe cofactors: R2lox and R2c proteins
Part of Journal of Biological Chemistry, p. 18372-18386, 2019
- DOI for Chemical flexibility of heterobimetallic Mn/Fe cofactors: R2lox and R2c proteins
- Download full text (pdf) of Chemical flexibility of heterobimetallic Mn/Fe cofactors: R2lox and R2c proteins
Part of mAbs, p. 1402-1414, 2019
- DOI for DuoMab: a novel CrossMab-based IgG-derived antibody format for enhanced antibody-dependent cell-mediated cytotoxicity
- Download full text (pdf) of DuoMab: a novel CrossMab-based IgG-derived antibody format for enhanced antibody-dependent cell-mediated cytotoxicity
Part of Biochimica et Biophysica Acta - Bioenergetics, 2019
Location-specific quantification of protein-bound metal ions by X-ray anomalous dispersion: Q-XAD
Part of Acta Crystallographica Section D, p. 764-771, 2019
- DOI for Location-specific quantification of protein-bound metal ions by X-ray anomalous dispersion: Q-XAD
- Download full text (pdf) of Location-specific quantification of protein-bound metal ions by X-ray anomalous dispersion: Q-XAD
Part of Journal of Biological Inorganic Chemistry, p. 849-861, 2019
- DOI for Redox-induced structural changes in the di-iron and di-manganese forms of Bacillus anthracis ribonucleotide reductase subunit NrdF suggest a mechanism for gating of radical access
- Download full text (pdf) of Redox-induced structural changes in the di-iron and di-manganese forms of Bacillus anthracis ribonucleotide reductase subunit NrdF suggest a mechanism for gating of radical access
Solving a new R2lox protein structure by microcrystal electron diffraction
Part of Science Advances, 2019
- DOI for Solving a new R2lox protein structure by microcrystal electron diffraction
- Download full text (pdf) of Solving a new R2lox protein structure by microcrystal electron diffraction
Part of Journal of the American Chemical Society, p. 1471-1480, 2018
- DOI for Driving Protein Conformational Changes with Light: Photoinduced Structural Rearrangement in a Heterobimetallic Oxidase
- Download full text (pdf) of Driving Protein Conformational Changes with Light: Photoinduced Structural Rearrangement in a Heterobimetallic Oxidase
Ether cross-link formation in the R2-like ligand-binding oxidase
Part of Journal of Biological Inorganic Chemistry, p. 879-886, 2018
- DOI for Ether cross-link formation in the R2-like ligand-binding oxidase
- Download full text (pdf) of Ether cross-link formation in the R2-like ligand-binding oxidase
Divergent assembly mechanisms of the manganese/iron cofactors in R2lox and R2c proteins
Part of Journal of Inorganic Biochemistry, p. 164-177, 2016
- DOI for Divergent assembly mechanisms of the manganese/iron cofactors in R2lox and R2c proteins
- Download full text (pdf) of Divergent assembly mechanisms of the manganese/iron cofactors in R2lox and R2c proteins
Part of Inorganic Chemistry, p. 9869-9885, 2016
Part of Journal of Physical Chemistry B, p. 13904-13921, 2015
- DOI for Characterization of Oxygen Bridged Manganese Model Complexes Using Multifrequency 17O-Hyperfine EPR Spectroscopies and Density Functional Theory
- Download full text (pdf) of Characterization of Oxygen Bridged Manganese Model Complexes Using Multifrequency 17O-Hyperfine EPR Spectroscopies and Density Functional Theory
Part of Nature Communications, 2015
- DOI for Crystal structure, biochemical and cellular activities demonstrate separate functions of MTH1 and MTH2
- Download full text (pdf) of Crystal structure, biochemical and cellular activities demonstrate separate functions of MTH1 and MTH2
Structural Basis for Oxygen Activation at a Heterodinuclear Manganese/Iron Cofactor
Part of Journal of Biological Chemistry, p. 25254-25272, 2015
- DOI for Structural Basis for Oxygen Activation at a Heterodinuclear Manganese/Iron Cofactor
- Download full text (pdf) of Structural Basis for Oxygen Activation at a Heterodinuclear Manganese/Iron Cofactor
Electronic Structural Flexibility of Heterobimetallic Mn/Fe Cofactors: R2lox and R2c Proteins
Part of Journal of the American Chemical Society, p. 13399-13409, 2014
- DOI for Electronic Structural Flexibility of Heterobimetallic Mn/Fe Cofactors: R2lox and R2c Proteins
- Download full text (pdf) of Electronic Structural Flexibility of Heterobimetallic Mn/Fe Cofactors: R2lox and R2c Proteins
Part of PLOS ONE, 2013
- DOI for Crystal Structure of an Anti-Ang2 CrossFab Demonstrates Complete Structural and Functional Integrity of the Variable Domain
- Download full text (pdf) of Crystal Structure of an Anti-Ang2 CrossFab Demonstrates Complete Structural and Functional Integrity of the Variable Domain
Part of Proceedings of the National Academy of Sciences of the United States of America, p. 17189-17194, 2013
- DOI for Direct observation of structurally encoded metal discrimination and ether bond formation in a heterodinuclear metalloprotein
- Download full text (pdf) of Direct observation of structurally encoded metal discrimination and ether bond formation in a heterodinuclear metalloprotein
Part of Metallomics, p. 894-898, 2012
- DOI for X-ray reduction correlates with soaking accessibility as judged from four non crystallographically related diiron sites
- Download full text (pdf) of X-ray reduction correlates with soaking accessibility as judged from four non crystallographically related diiron sites
Structure and DNA-binding activity of the Pyrococcus furiosus SMC protein hinge domain
Part of Proteins, p. 558-568, 2011
- DOI for Structure and DNA-binding activity of the Pyrococcus furiosus SMC protein hinge domain
- Download full text (pdf) of Structure and DNA-binding activity of the Pyrococcus furiosus SMC protein hinge domain
Part of Nucleic Acids Research, p. 3454-3465, 2010
- DOI for Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins
- Download full text (pdf) of Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins
Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase
Part of Nature, p. 1015-1018, 2009
Part of Chemical Physics, p. 130-141, 2008
Part of Journal of Molecular Biology, p. 140-152, 2006
People
Julia Griese, Assistant Professor, Principal Investigator
email: julia.griese@icm.uu.se
phone: +46-18-471 4982
Oksana Koshla, PhD, Postdoc
Former members
- Linda Juniar, PhD, Postdoc
- Shruthi Krishnaswamy, Project student
- Stijn Tieleman, Bachelor student
- Deniz Biçer, Project student
- Emmanuel Nji, PhD, Visiting Researcher
- Carita Hammar Emas, Master student spring 2022
- Yozlem Dilaver, Project student summer 2021
- Francisco Javier Marcos-Torres, PhD, Postdoc 2018-2020
- Dirk Maurer, PhD, Researcher 2018-2020
- Mangirdas Kežys, Master student spring 2020
- Malin Svensson, Master student spring 2020
- Grim Elison Kalman, Master student spring 2019
- Dilay Yılmaz, Project student summer 2018

