Rosling lab: Our research
Ecology and evolution of symbiotic fungi
Under natural conditions the health and survival of plants depend on their symbiotic fungal partners. Molecular identification methods have revolutionized soil microbial ecology and unraveled a tremendous fungal diversity in soil that we are only beginning to truly appreciate. Many of the most abundant organisms recovered from soil fungal metabarcoding studies remain unknown and one of the long-term goal of our research is to recognize understand some of the abundant yet unknown members of soil fungal communities.
Our research focuses primarily on arbuscular mycorrhizal fungi and the class Archaeorhizomycetes but we are broadly interested in hidden fungal diversity and symbiotic interactions. Topics that has developed into interesting collaborations on mycobiont and photobiont diversity of lichens.
Evolutionary history of AM fungi
Between 2016-2022, funding from ERC allowed us to develop a new research program on arbuscular mycorrhizal (AM) fungi (HeteroDynamic). AM fungi form symbiotic interactions with almost all terrestrial plants and have done so since plants first colonized land. We are interested in their diversity, evolutionary history and genome organization. The research program involved genome sequencing and culture experiments and aims at characterizing the structure of genetic variability within AM fungal species and is a collaboration with Prof James Bever at the University of Kansas. Two PhD thesis has been completed within the research program. Dr. Shadi Eshghi Saraei in 2022 and Dr. Mercè Montoliu Nerin in 2020. PhD student David Manyara continues the work to understand evolutionary histories of AM fungi.
%20c_405796-l_1-k_kungsa-ngslilja.jpg)
Fritillaria meleagris in Kungsängen, Uppsala. Photo: Anna Rosling
Diversity and ecology of Archaeorhizomycetes
We combine eDNA metabarcoding with fungal culturing efforts to understand and describe the unknown majority of fungi that live in our soils. This approach has allowed us to describe the ubiquitous ancient fungal class Archaeorhizomycetes, a.k.a. Soil Clone Group 1, (Science 333, 876-879).
This fungal class is diverse and occur in roots and soil from most terrestrial ecosystems but their ecological roles and means of dispersal remains unknown. In our research we continue to explore function, distribution and diversity of the class. Efforts to capture more species in culture are mostly fruitless and we argue that eDNA sequencing data can actually provide strong evidence for delaminating species in the class (Kalsoom Khan et al 2020). One PhD thesis has been completed within the research program, Dr. Kerri Kluting in 2022. Dr Veera Tuovinen joined the group as a post doc in 2019 and continues the work to describe unknown species of Archaeorhizomycetes.
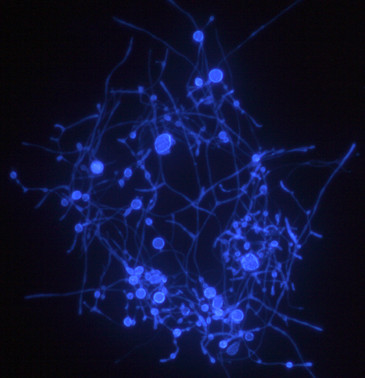
Calcoflour stained mycelia of A. finlayi. Photo: Audrius Menkis
Fungal activity and soil nutrient availability
Soils develop as a result of their biological processes while constrained by the environmental and geochemical conditions of a site. Soils in turn provide the environmental constrains that soil fungi adapt to, in order to, form associations with host, compete for substrate and to complete their life cycle. Fungi are well adapted to the temporally and spatially heterogeneous soil environment. Thin hyphae allow for efficient substrate exploration and provide large surface area for uptake. Resource translocation within the mycelia allows fungi to link spatially separated substrates, i.e. the carbon supply in its host roots with sources for nutrient uptake in soil. Through collaborations with chemists and geochemists the group contributes to research on soil organic matter decomposition and enhanced silica weathering as a possible mean for carbon sequestration.

Pine forest, Jädraås, Sweden. Photo: Anna Rosling
Resources for the Rosling lab
Using long read rDNA amplicon sequencing we characterised the soil fungal community across three soil horizons at the Ivantjärnsheden field station close to Jädraås (60°49′N, 16°30′E, altitude 185 m), Sweden (published in IMA Fungus). The complete “phylogenetic” dataset with 272 fungal ASVs is available via the GBIF repository.
The annotation pipeline used to annotate genome assemblies of Claroideoglomus claroideum/C. luteum (SA101) in "Building de novo reference genome assemblies of complex eukaryotic microorganisms from single nuclei” by Montoliu-Nerin 2020 is available via bitbucket.
