The MYC Oncoprotein
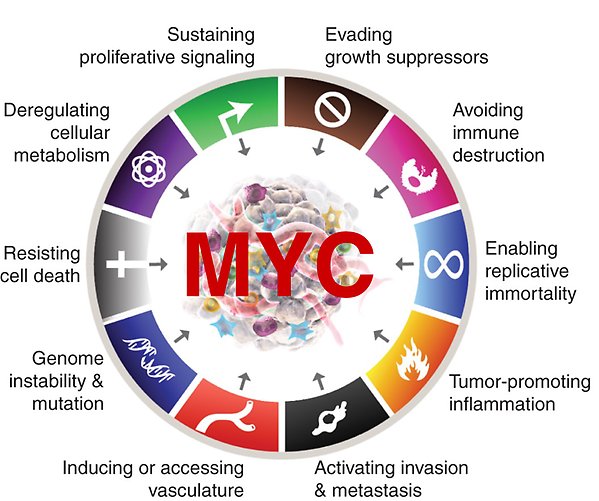
Hallmarks
Description
The Lars-Gunnar Larsson group’s research focuses on studies of the function, regulation and targeting of the MYC oncoprotein. The MYC oncogene family encodes transcription factors that regulate a vast number of genes involved in cell growth, apoptosis, metabolism, differentiation, senescence, stemness, immune responses and other cellular functions. Deregulated expression of MYC contributes to the development of over half of all human cancers, and often correlates with advanced stages of disease, resistance to therapy and poor prognosis. Our research addresses questions on how MYC cooperates and interacts with other factors to regulate gene transcription and how this affects biological processes of relevance for tumor development, such as senescence suppression and immune surveillance. We are also interested in how MYC’s function, activity and stability is regulated through interaction with such factors. Finally, we are involved in a campaign to identify and validate molecules that target MYC directly or indirectly with the aim to develop new cancer therapies.
Selected publications
MYCN Amplification Is Associated with Reduced Expression of Genes Encoding γ-Secretase Complex and NOTCH Signaling Components in Neuroblastoma.
Agarwal P, Glowacka A, Mahmoud L, Bazzar W, Larsson LG, Alzrigat M
Int J Mol Sci 2023 May;24(9):
A MYC-controlled redox switch protects B lymphoma cells from EGR1-dependent apoptosis.
Yao H, Chen X, Wang T, Kashif M, Qiao X, Tüksammel E, Larsson LG, Okret S, Sayin VI, Qian H, Bergo MO
Cell Rep 2023 Aug;42(8):112961
MYCMI-7: A Small MYC-Binding Compound that Inhibits MYC: MAX Interaction and Tumor Growth in a MYC-Dependent Manner.
Castell A, Yan Q, Fawkner K, Bazzar W, Zhang F, Wickström M, Alzrigat M, Franco M, Krona C, Cameron DP, Dyberg C, Olsen TK, Verschut V, Schmidt L, Lim SY, Mahmoud L, Hydbring P, Lehmann S, Baranello L, Nelander S, Johnsen JI, Larsson LG
Cancer Res Commun 2022 Mar;2(3):182-201
MYC Inhibition Halts Metastatic Breast Cancer Progression by Blocking Growth, Invasion, and Seeding.
Massó-Vallés D, Beaulieu ME, Jauset T, Giuntini F, Zacarías-Fluck MF, Foradada L, Martínez-Martín S, Serrano E, Martín-Fernández G, Casacuberta-Serra S, Castillo Cano V, Kaur J, López-Estévez S, Morcillo MÁ, Alzrigat M, Mahmoud L, Luque-García A, Escorihuela M, Guzman M, Arribas J, Serra V, Larsson LG, Whitfield JR, Soucek L
Cancer Res Commun 2022 Feb;2(2):110-130
HIF-1 stabilization in T cells hampers the control of Mycobacterium tuberculosis infection.
Liu R, Muliadi V, Mou W, Li H, Yuan J, Holmberg J, Chambers BJ, Ullah N, Wurth J, Alzrigat M, Schlisio S, Carow B, Larsson LG, Rottenberg ME
Nat Commun 2022 Sep;13(1):5093
Methods to Study Myc-Regulated Cellular Senescence: An Update.
Zhang F, Bazzar W, Alzrigat M, Larsson LG
Methods Mol Biol 2021 ;2318():241-254
The novel low molecular weight MYC antagonist MYCMI-6 inhibits proliferation and induces apoptosis in breast cancer cells. AlSultan D, Kavanagh E, O'Grady S, Eustace AJ, Castell A, Larsson LG, Crown J, Madden SF, Duffy MJ Invest New Drugs 2021 04;39(2):587-594
Pharmacological inactivation of CDK2 inhibits MYC/BCL-XL-driven leukemia in vivo through induction of cellular senescence. Bazzar W, Bocci M, Hejll E, Högqvist Tabor V, Hydbring P, Grandien A, Alzrigat M, Larsson LG Cell Cycle 2021 01;20(1):23-38
PTEN and DNA-PK determine sensitivity and recovery in response to WEE1 inhibition in human breast cancer. Brunner A, Suryo Rahmanto A, Johansson H, Franco M, Viiliäinen J, Gazi M, Frings O, Fredlund E, Spruck C, Lehtiö J, Rantala JK, Larsson LG, Sangfelt O Elife 2020 07;9():
A selective high affinity MYC-binding compound inhibits MYC:MAX interaction and MYC-dependent tumor cell proliferation. Castell A, Yan Q, Fawkner K, Hydbring P, Zhang F, Verschut V, Franco M, Zakaria SM, Bazzar W, Goodwin J, Zinzalla G, Larsson LG Sci Rep 2018 07;8(1):10064
MYC Modulation around the CDK2/p27/SKP2 Axis.
Hydbring P, Castell A, Larsson LG
Genes (Basel) 2017 Jun;8(7):
Interferon-γ-induced p27KIP1 binds to and targets MYC for proteasome-mediated degradation.
Bahram F, Hydbring P, Tronnersjö S, Zakaria SM, Frings O, Fahlén S, et al
Oncotarget 2016 Jan;7(3):2837-54
Targeting MYC Translation in Colorectal Cancer.
Castell A, Larsson LG
Cancer Discov 2015 Jul;5(7):701-3
MYC synergizes with activated BRAFV600E in mouse lung tumor development by suppressing senescence.
Tabor V, Bocci M, Alikhani N, Kuiper R, Larsson LG
Cancer Res. 2014 Aug;74(16):4222-9
CDK-mediated activation of the SCF(FBXO) (28) ubiquitin ligase promotes MYC-driven transcription and tumourigenesis and predicts poor survival in breast cancer.
Cepeda D, Ng HF, Sharifi HR, Mahmoudi S, Cerrato VS, Fredlund E, Magnusson K, Nilsson H, Malyukova A, Rantala J, Klevebring D, Viñals F, Bhaskaran N, Zakaria SM, Rahmanto AS, Grotegut S, Nielsen ML, Szigyarto CA, Sun D, Lerner M, Navani S, Widschwendter M, Uhlén M, Jirström K, Pontén F, Wohlschlegel J, Grandér D, Spruck C, Larsson LG, Sangfelt O
EMBO Mol Med 2013 Jul;5(7):1067-86
The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop.
Menssen A, Hydbring P, Kapelle K, Vervoorts J, Diebold J, Lüscher B, et al
Proc. Natl. Acad. Sci. U.S.A. 2012 Jan;109(4):E187-96
Oncogene- and tumor suppressor gene-mediated suppression of cellular senescence.
Larsson LG
Semin. Cancer Biol. 2011 Dec;21(6):367-76
Tipping the balance: Cdk2 enables Myc to suppress senescence.
Hydbring P, Larsson LG
Cancer Res. 2010 Sep;70(17):6687-91
The Yin and Yang functions of the Myc oncoprotein in cancer development and as targets for therapy.
Larsson LG, Henriksson MA
Exp. Cell Res. 2010 May;316(8):1429-37
Cdk2 suppresses cellular senescence induced by the c-myc oncogene.
Campaner S, Doni M, Hydbring P, Verrecchia A, Bianchi L, Sardella D, et al
Nat. Cell Biol. 2010 Jan;12(1):54-9; sup pp 1-14
Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation.
Hydbring P, Bahram F, Su Y, Tronnersjö S, Högstrand K, von der Lehr N, et al
Proc. Natl. Acad. Sci. U.S.A. 2010 Jan;107(1):58-63
Direct observation of individual endogenous protein complexes in situ by proximity ligation.
Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius KJ, Jarvius J, et al
Nat. Methods 2006 Dec;3(12):995-1000
c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription.
Arabi A, Wu S, Ridderstråle K, Bierhoff H, Shiue C, Fatyol K, et al
Nat. Cell Biol. 2005 Mar;7(3):303-10
The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription.
von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, et al
Mol. Cell 2003 May;11(5):1189-200
c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover.
Bahram F, von der Lehr N, Cetinkaya C, Larsson LG
Blood 2000 Mar;95(6):2104-10
Doctoral dissertations from the group
Doktorsavhandlingar
2001
Siqin Wu. “The Myc network of growth regulators and its interplay with growth and differentiation signals”
https://uu.diva-portal.org/smash/record.jsf?pid=diva2:160820
Anne Hultquist. ”Regulation and function of the Mad/Max/Myc network during neuronal and hematopoietic differentiation”
https://uu.diva-portal.org/smash/record.jsf?pid=diva2:168566
2003
Natalie von der Lehr. ”The transcriptional function of the c-Myc oncoprotein and its regulation by the ubiquitin/proteasome pathway”.
https://res.slu.se/id/publ/11408
2004
Fuad Bahram. ” Post-translational regulation of Myc oncoprotein function”.
https://res.slu.se/id/publ/12156
2005
Cihan Cetinkaya. “Control of growth and differentiation of human neuronal and hematopoietic tumour cells via Myc/Max/Mad-network proteins”.
https://res.slu.se/id/publ/12593
2008
Su Yingtao. ”Function and regulation of myc-family bHLHZip transcription factors during the animal and plant cell cycle”.
https://res.slu.se/id/publ/18012
Sara Fahlén. ”Regulation of Myc oncoprotein function by E3 ubiquitin ligases”.
https://res.slu.se/id/publ/19569
2009
Per Hydbring. ”Modulating the activity of the c-Myc oncoprotein : implications for therapeutic treatment”.
https://res.slu.se/id/publ/22771
Karin Ridderstråle. ”Myc oncoprotein cofactor interactions: function, regulation and targeting”.
http://hdl.handle.net/10616/39857
2012
Eduar Hejll. ”Vascular development and safeguard mechanisms against tumorigenesis : oncogene-induced apoptosis and cellular senescence”.
http://hdl.handle.net/10616/41158
2013
Hamid Reza Sharifi. “Interplay between the Myc oncoprotein, cyclin-dependent kinases and E3 ubiquitin ligases”
http://hdl.handle.net/10616/41813
2019
Qinzi Yan. “Identification and characterization of small molecules targeting MYC function”.
http://hdl.handle.net/10616/46785
Siti Mariam Zakaria. ”Cooperativity between MYC and other oncogenic factors : implications for tumorigenesis and targeting of MYC”.
http://hdl.handle.net/10616/46630
Fan Zhang. ”Pro-senescence therapy by targeting MYC and its network as a strategy to combat cancer”.
http://hdl.handle.net/10616/46642
2021
Wesam Bazzar. ”Targeting MYC and CDK2 in cancer : impacts on senescence, apoptosis and immune modulation”.
