Holmqvist lab
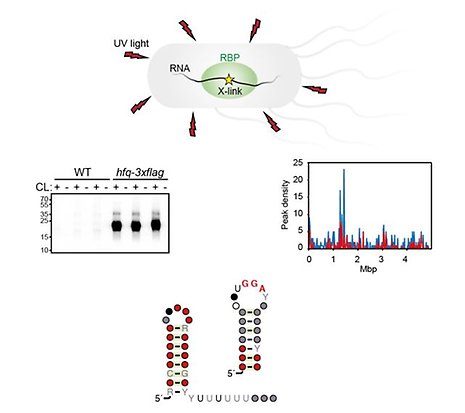
Bacteria are constantly challenged by harsh conditions in their ever-changing natural environments. To survive and proliferate, bacterial cells cope with stressful conditions by rapidly re-wire gene expression. To this end, bacteria have acquired complex gene regulatory networks, which at the post-transcriptional level are dominated by RNA-binding proteins and regulatory small RNAs. We study molecular mechanisms and cellular functions of bacterial RNA-binding proteins and their RNA ligands to understand how these regulatory macromolecules contribute to bacterial growth and survival.
Popular science presentation
...
Research projects
The aim of our research is to understand biological functions and regulatory mechanisms of RNA-binding proteins (RBPs) and regulatory RNAs in pathogenic bacteria. This will lead to a better understanding of the regulatory decisions made by bacteria in adverse environments, such as inside the infected host.
RBPs are built of RNA-binding domains that recognize distinct motifs buried in RNA transcripts. The specific interactions between RBPs and their RNA ligands often result in regulation of gene expression, e.g. by activating or inhibiting mRNA translation, or by altering RNA stability. To understand the function of any given RBP, it is key to identify both its RNA ligands and the specific binding motifs. To do this in a comprehensive manner, we use CLIP-seq (crosslinking and immunoprecipitation sequencing) that allows for simultaneous identification of all RNA sequences bound by an RBP at any given moment. This approach can faithfully inform on cellular recognition motifs and RBP specificities, and reveal regulated cellular processes and regulatory mechanisms. We study Salmonella as a model system for bacterial pathogenesis. Salmonella causes typhoid fever and gastroenteritis leading annually to hundreds of thousands of deaths. Within the human host, a number of harsh environments force bacterial pathogens to rapidly change their physiological state by re-wiring gene expression and activate virulence gene expression programs. To fully understand the infection process, it is therefore critical to understand how gene expression is controlled within the pathogen.
Group members
Publications
Part of mSphere, 2024
- DOI for ProQ-dependent activation of Salmonella virulence genes mediated by post-transcriptional control of PhoP synthesis
- Download full text (pdf) of ProQ-dependent activation of Salmonella virulence genes mediated by post-transcriptional control of PhoP synthesis
Rescue of Escherichia coli auxotrophy by de novo small proteins
Part of eLIFE, 2023
- DOI for Rescue of Escherichia coli auxotrophy by de novo small proteins
- Download full text (pdf) of Rescue of Escherichia coli auxotrophy by de novo small proteins
RNA interactome capture in Escherichia coli globally identifies RNA-binding proteins
Part of Nucleic Acids Research, p. 4572-4587, 2023
- DOI for RNA interactome capture in Escherichia coli globally identifies RNA-binding proteins
- Download full text (pdf) of RNA interactome capture in Escherichia coli globally identifies RNA-binding proteins
CsrA enters Hfq's territory: Regulation of a base-pairing small RNA
Part of Molecular Microbiology, p. 4-9, 2022
- DOI for CsrA enters Hfq's territory: Regulation of a base-pairing small RNA
- Download full text (pdf) of CsrA enters Hfq's territory: Regulation of a base-pairing small RNA
The RNA-binding protein ProQ promotes antibiotic persistence in Salmonella
Part of Molecular and Cellular Biology, 2022
- DOI for The RNA-binding protein ProQ promotes antibiotic persistence in Salmonella
- Download full text (pdf) of The RNA-binding protein ProQ promotes antibiotic persistence in Salmonella
Part of Nucleic Acids Research, p. 9992-10006, 2021
- DOI for Saturation mutagenesis charts the functional landscape of Salmonella ProQ and reveals a gene regulatory function of its C-terminal domain
- Download full text (pdf) of Saturation mutagenesis charts the functional landscape of Salmonella ProQ and reveals a gene regulatory function of its C-terminal domain
Part of RNA Biology, p. 872-880, 2020
- DOI for Small RNAs OmrA and OmrB promote class III flagellar gene expression by inhibiting the synthesis of anti-Sigma factor FlgM
- Download full text (pdf) of Small RNAs OmrA and OmrB promote class III flagellar gene expression by inhibiting the synthesis of anti-Sigma factor FlgM
The Length of a DNA T-Tract Modulates Expression of a Virulence-Regulating sRNA
Part of Molecular Cell, p. 175-177, 2020
Part of mBio, 2020
- DOI for The Small Toxic Salmonella Protein TimP Targets the Cytoplasmic Membrane and Is Repressed by the Small RNA TimR
- Download full text (pdf) of The Small Toxic Salmonella Protein TimP Targets the Cytoplasmic Membrane and Is Repressed by the Small RNA TimR
Hfq-dependent mRNA unfolding promotes sRNA-based inhibition of translation
Part of EMBO Journal, 2019
- DOI for Hfq-dependent mRNA unfolding promotes sRNA-based inhibition of translation
- Download full text (pdf) of Hfq-dependent mRNA unfolding promotes sRNA-based inhibition of translation
Part of Molecular Cell, p. 971-982, 2018
Structure of the Escherichia coli ProQ RNA-binding protein
Part of RNA, p. 696-711, 2017
- DOI for Structure of the Escherichia coli ProQ RNA-binding protein
- Download full text (pdf) of Structure of the Escherichia coli ProQ RNA-binding protein
Part of Nucleic Acids Research, 2013
- DOI for Massive functional mapping of a 5'-UTR by saturation mutagenesis, phenotypic sorting and deep sequencing
- Download full text (pdf) of Massive functional mapping of a 5'-UTR by saturation mutagenesis, phenotypic sorting and deep sequencing
A mixed double negative feedback loop between the sRNA MicF and the global regulator Lrp
Part of Molecular Microbiology, p. 414-427, 2012
Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis
Part of EMBO Journal, p. 1840-1850, 2010
Part of PLoS Genetics, 2009
Hfq-dependent regulation of OmpA synthesis is mediated by an an-tisense RNA.
Part of Genes & Development, p. 2355-2366, 2005
People
Erik Holmqvist, PhD
Group leader
Sofia Berggren, PhD student
Thomas Stenum, Postdoc
Athina Eleftheraki, PhD student
Sophie Baars, Master thesis student
Margherita Panza, Master thesis erasmus student
Group alumni
Ankith Kumar, Master thesis student
Mikael Strandgren, Master thesis student
Liis Andresen, Postdoc
Zeynep Iloglu, Erasmus student
Jennifer Schulz, Master thesis student
Dagny Vilhelmsdottir, Master thesis student
Joel Striem, Master thesis student
Oskar Bergman, Master thesis student
Viktor Björklund, Master thesis student
Josefin Nilsson Zangelin, Master thesis student
Yolanda Martinez Burgo, Postdoc

