Antibiotic resistance by target binding, an unexplored mechanism of clinical relevance
Antibiotic resistance is commonly mediated by efflux pumps, mutations or modifications of the target, degradation or modification of the drug, or factor-assisted protection. Of these, factor-assisted protection mechanisms are the least understood. They involve the expression of a protein that interacts with the target of the antibiotic and provide resistance without direct interaction with the drug. In this project, we will study one of these systems: FusB-mediated resistance to fusidic acid.
Fusidic acid is an antibiotic effective mainly against gram positive bacteria and it is commonly used against Staphylococcus aureus infections. Fusidic acid stalls protein synthesis by binding to elongation factor G (EF-G) after translocation and prevents its release from the ribosome. This, in turn, hinders bacterial growth. Unfortunately, resistant strains reduce its efficacy. There are multiple fusidic acid resistance types: FusA, FusB/C/D/F (FusB-type), and FusE. FusA and FusE constitute mutations of the target; FusA on EF-G and FusE on ribosomal protein L6, which is positioned in the region where EF-G binds to the ribosome. On the other hand, the FusB type functions through factor-assisted protection and it is the most prevalent in many clinical isolates.
FusB-type resistance to fusidic acid is mediated by the expression of FusB, a small protein that binds to EF-G, inducing resistance. They form a tight complex and there is evidence that FusB causes increased turnover of EF-G in the ribosome.
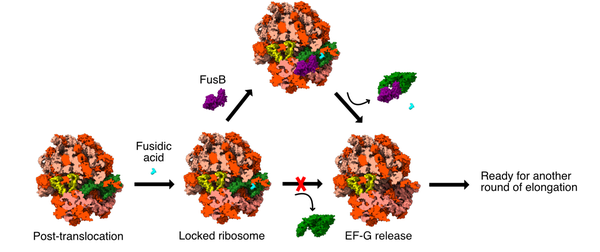
Schematic of fusidic acid resistance mediated by FusB. Fusidic acid (cyan) binds to EF-G (green) in the ribosome after translocation, inhibiting its release. FusB (purple) binds to EF-G and rescues EF-G from the ribosome to start another elongation cycle.
The structures of EF-G and FusB from S. aureus have been solved by our group and there is also an NMR (nuclear magnetic resonance) structure of a truncated FusB-EF-G complex (lacking domains I-II of EF-G). In this NMR study they observed increased dynamics of domain III of EF-G in the complex, indicating that this effect could play an important role in the mechanism of resistance. However, many aspects of the mechanism are still unclear.
Understanding this mechanism of resistance is of great importance to find new ways to counteract resistant strains and in addition will increase our knowledge of translation and EF-G function in the cell. Furthermore, other uncharacterized resistance mechanisms might have similarities that could be identified.
To accomplish the goals of this project, we will employ structural biology techniques such as cryo-EM, X-ray crystallography and small-angle X-ray scattering to generate hypotheses that we can further test using biochemistry and microbiology.
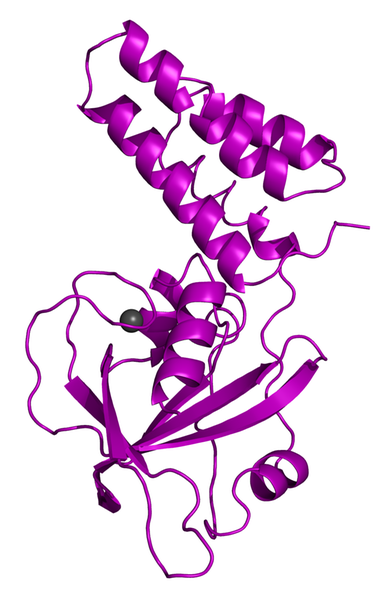
X-ray crystal structure of FusB.
Related published research
- Chen, Yang, Ravi Kiran Koripella, Suparna Sanyal, and Maria Selmer. 2010. ‘Staphylococcus Aureus Elongation Factor G - Structure and Analysis of a Target for Fusidic Acid: Crystal Structure of Staphylococcus Aureus EF-G’. FEBS Journal 277 (18): 3789–3803
- Cox, G., G. S. Thompson, H. T. Jenkins, F. Peske, A. Savelsbergh, M. V. Rodnina, W. Wintermeyer, S. W. Homans, T. A. Edwards, and A. J. O’Neill. 2012. ‘Ribosome Clearance by FusB-Type Proteins Mediates Resistance to the Antibiotic Fusidic Acid’. Proceedings of the National Academy of Sciences 109 (6): 2102–7
- Cox, Georgina, Thomas A. Edwards, and Alex J. O’Neill. 2013. ‘Mutagenesis Mapping of the Protein-Protein Interaction Underlying FusB-Type Fusidic Acid Resistance’. Antimicrobial Agents and Chemotherapy 57 (10): 4640–44
- Guo, Xiaohu, Kristin Peisker, Kristina Bäckbro, Yang Chen, Ravi Kiran Koripella, Chandra Sekhar Mandava, Suparna Sanyal, and Maria Selmer. 2012. ‘Structure and Function of FusB: An Elongation Factor G-Binding Fusidic Acid Resistance Protein Active in Ribosomal Translocation and Recycling’. Open Biology 2 (3): 120016.
- O’Neill, A. J., F. McLaws, G. Kahlmeter, A. S. Henriksen, and I. Chopra. 2007. ‘Genetic Basis of Resistance to Fusidic Acid in Staphylococci’. Antimicrobial Agents and Chemotherapy 51 (5): 1737–40.
- O’Neill, Alexander John, and Ian Chopra. 2006. ‘Molecular Basis of FusB -Mediated Resistance to Fusidic Acid in Staphylococcus Aureus: Fusidic Acid Resistance in S. Aureus’. Molecular Microbiology 59 (2): 664–76.
- Tomlinson, Jennifer H., Arnout P. Kalverda, and Antonio N. Calabrese. 2020. ‘Fusidic Acid Resistance through Changes in the Dynamics of the Drug Target’. Proceedings of the National Academy of Sciences 117 (41): 25523–31
- Tomlinson, Jennifer H., Gary S. Thompson, Arnout P. Kalverda, Anastasia Zhuravleva, and Alex J. O’Neill. 2016. ‘A Target-Protection Mechanism of Antibiotic Resistance at Atomic Resolution: Insights into FusB-Type Fusidic Acid Resistance’. Scientific Reports 6 (1): 19524
