Diagnostics with magnetic nanoparticles
The overall aim is to develop a sensitive, low-cost and easy-to-use diagnostic method, which can serve as an efficient analytical platform for rapid detection and characterization of antibiotic-resistant bacteria.
The detection assay combines magnetic detection of changes in Brownian rotation dynamics of functionalized magnetic nanoparticles with the padlock- probe-ligation technique and rolling circle amplification of the probe-target DNA complex. An increased in hydrodynamic volume caused by binding of target molecules to the probes on the surface of the nanoparticles, causes a larger rotation time, and thus a lower rotation frequency. By detecting the frequency shift in the peak position of the imaginary part of the magnetic susceptibility the number of attached target molecules can be probed.
The project aims to study, adapt and optimize the magnetic biosensing method to make it able to detect and analyse relevant pathogens resistant to antibiotics. New probe molecules will be designed and the procedure will be optimized with regard to detection speed, sensibility and usability.
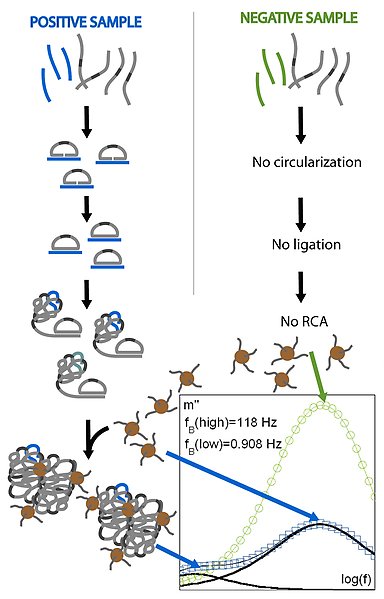
Schematic illustration of the Volume-Amplified Magnetic Nanobeads Detection Assay (VAM-NDA). After addition of padlock probes (grey) to the target DNA molecules (blue), circularized padlock probes are formed by ligation. The circularized molecules are then amplified by the RCA mechanism, creating DNA macromolecules. To detect the presence of the DNA macromolecules, complementary DNA-tagged magnetic nanoparticles (brown) are added, which are bound to the DNA macromolecules. The particle incorporation results in a spectrum of the complex magnetization (positive sample, blue magnetization curve) substantially different from the response of a sample containing no target DNA and only free nanoparticles (negative sample, green magnetization curve).
Related published research
- Homogeneous differential magnetic assay. S. Sepehri, T. Zardán Gómez de la Torre, J. F. Schneiderman, A. Jesorka, C. Johansson, M. Nilsson, J. Albert, M. Strømme, D. Winkler, A. Kalaboukhov ACS Sensors (2019)
- A magnetic bead-based read-out procedure for rapid detection of DNA molecules. T. Zardán Gómez de la Torre, D. Herthnek and M. Strømme Journal of Nanoscience and Nanotechnology 17 (2017) 2861-2864
- Novel readout method for molecular diagnostic assays based on optical measurements of magnetic nanobead dynamics. M. Donolato, P. Antunes, R. S. Bejhed, T. Zardán Gómez de la Torre, F. W. Østerberg, M. Strömberg, M. Nilsson, M. Strømme, P. Svedlindh, M. F. Hansen and P. Vavassori Analytical Chemistry 87 (2015) 1622–1629
- Optomagnetic read-out enables easy, rapid, and cost-efficient qualitative biplex detection of bacterial DNA sequences. R. S. Bejhed, T. Zardán Gómez de la Torre, P. Svedlindh and M. Strömberg Biotechnology Journal 10 (2015) 469–472
- A magnetic nanobead-based bioassay provides sensitive detection of single- and biplex bacterial DNA using a portable AC susceptometer. M. Strömberg, T. Zardán Gómez de la Torre, M. Nilsson, P. Svedlindh and M. Strømme Biotechnology Journal 9 (2014) 137–145
- Detection of rifampicin resistance in Mycobacterium tuberculosis by padlock probes and magnetic nanobead-based readout. A. Engström, T. Zardán Gómez de la Torre, M. Strømme, M. Nilsson and D. Herthnek PLOS ONE 8 (2013) e62015
