Formation of Persister Bacteria in Salmonella and the Role of ProQ
The discovery of antibiotics has revolutionized the field of medicine, saving countless of lives. However, with time, bacteria have developed different strategies to survive antibiotic treatment. The emergence of antibiotic resistance is the predominant one, referring to the ability of a bacterium, to grow in the presence of an antibiotic. In addition to antibiotic resistance, the concept of bacterial persistence is gaining attention as an alternative strategy for bacterial survival in the presence of antibiotics.
Bacterial persistence allows a subpopulation of bacteria to temporarily withstand antibiotic treatment by entering a dormant-like state. In this state, persister bacteria slow down growth and keep a low metabolic activity. This persister lifestyle protects bacteria from antibiotic killing, since the efficacy of antibiotics generally requires metabolic activity. Once the antibiotic is removed, persister bacteria can return to an active lifestyle, reproduce and form a new bacterial population. Increasing evidence suggests that these persister bacteria can cause prolonged and relapsed bacterial infections, and promote the emergence of resistant bacteria, rendering antibiotic treatment ineffective.
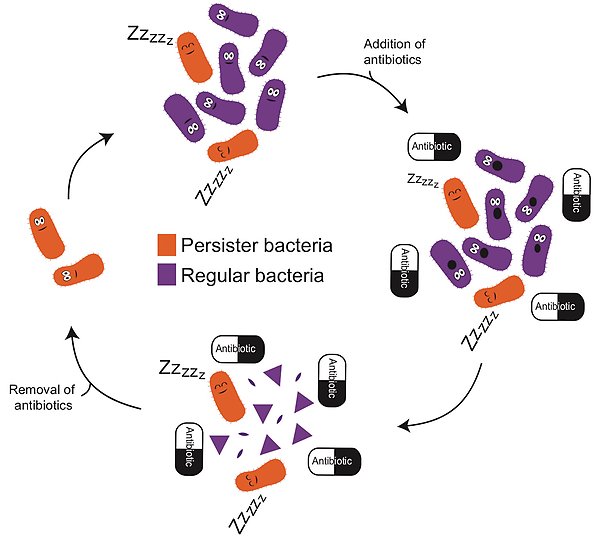
How persister bacteria and regular bacteria react to antibiotic treatment
nfections caused by the pathogenic bacterium Salmonella are a major challenge in both human and animal health. Salmonella is the cause of over one billion infections annually, typically resulting in gastroenteritis, and typhoid fever. Antibiotics are routinely used for treating infections caused by Salmonella. Recently, it was discovered that upon infection of host cells, Salmonella can form a large fraction of persister bacteria which survive antibiotic exposure. However, the mechanism behind Salmonella persister formation remains unclear. Understanding mechanisms of action could help us to prevent and control bacterial persistence, and thereby improve the outcome of Salmonella infections.
In this UAC project, we are studying the formation of persister bacteria in Salmonella with focus on the protein ProQ. ProQ is a global RNA-binding and regulatory protein, which associates with a large repertoire of RNA molecules in the bacterial cell. The association of ProQ with RNA molecules leads to changes in gene expression and modulates the lifestyle of bacteria. It was previously shown that ProQ binds RNA molecules that are known to be involved in persister formation. We therefore hypothesized an important role for ProQ in bacterial persistence. We discovered that ProQ is indeed involved in the persister phenotype in Salmonella, and promotes the formation of persister bacteria. ProQ was found to aid in persister formation during exposure to different classes of antibiotics, and during various conditions, most importantly host cell infections. This was true for different Salmonella strains tested in our experiments. Together, our results show that ProQ has a key role in persister formation, and helps Salmonella to survive antibiotic treatment. The next step in our research is to investigate the molecular mechanisms behind the effect of ProQ on persister formation in Salmonella. A better understanding of this will help us to figure out how we can stop persister bacteria to form and, in doing so, improve antibiotic treatment.
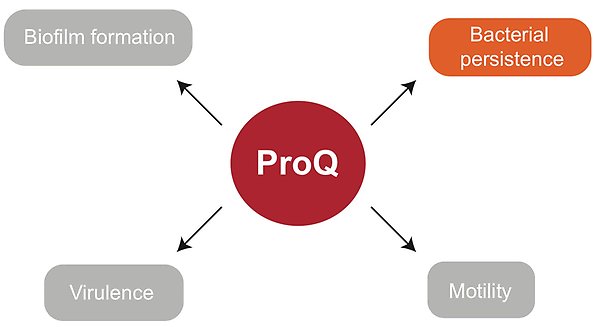
How ProQ influences in bacterial physiology
Related published research
- Alisa Rizvanovic, Jonas Kjellin, Fredrik Söderbom, Erik Holmqvist. Saturation mutagenesis charts the functional landscape of Salmonella ProQ and reveals a gene regulatory function of its C-terminal domain. 2021. Nucleic Acids Research.
- Balaban Q. Nathalie, Helaine Sophie, Lewis Kim, Ackermann Martin, Aldridge Bree, Andersson A. Dan, Brynildsen P. Mark, Bumann Dirk, Camilli Andrew, Collins J. James, Dehio Christoph, Fortune Sarah, Ghigo Jean-Marc, Hardt Wolf-Dietrich, Harms Alexander, Heinemann Matthias, Hung T. Deborah, Jenal Urs, Levin R. Bruce, Michiels Jan, Storz Gisela, Tan Man-Wan, Tenson Tanel, Van Melderen Laurence, Zinkernagel Annelies. 2019. “Definitions and guidelines for research on antibiotic persistence”. Nature Review Microbiology, no. 17 (July/2019): 441-448.
- Fisher A. Robert, Gollan Bridget, and Helaine Sophie. 2017. “Persistent bacterial infections and persister cells”. Nature Reviews Microbiology, no. 15 (August): 453-464.
- Gonzales M. Grecis, Hardwick W. Steven, Maslen L. Sarah, Skehel Mark, Holmqvist Erik, Vogel Jörg, Bateman Alex, Luisi F. Ben, Broadhurst R. William. 2017. “Structure of the Escherichia coli ProQ RNA-binding protein”. RNA, no. 23 (February): 696-711.
- Helaine Sophie, Cheverton Angela, Watson G. Kathryn, Matthews Sophie, Faure Laura, and Holden W. David. 2014. “Internalisation of Salmonella by macrophages induces formation of non-replicating persisters”. Science, no. 343 (6167): 204-208.
- Holmqvist Erik, Bergman Sofia, and Rizvanovic Alisa. In press. ”RNA-binding activity and regulatory functions of the emerging sRNA-binding protein ProQ”. BBA Gene Regulatory Mechanisms, no. 1863 (9).
- Holmqvist Erik, Li Lei, Bischler Thorsten, Barquist Lars, Vogel Jörg. 2018. ”Global Maps of ProQ Binding In vivo Reveal Target Recognition via RNA structure and Stability Control at mRNA 3’Ends”. Molecular Cell, no. 70 (5): 971-982.
- Holmqvist Erik, and Vogel Jörg. 2019. ”RNA-binding proteins in bacteria”. Nature Reviews Microbiology, no 16 (October/2018): 601-615.
- Smirnov Alexandre, Förstner U. Konrad, Holmqvist Erik, Otto Andreas, Gunster Regina, Becher Dörte, Reinhardt Richard, and Vogel Jörg. 2016. ”Grad-seq guides the discovery of ProQ as a major small RNA-binding protein”. PNAS, no. 113 (41/October): 11591-11596.
