Chromosomal Hybrids: A source of globally-spread antibiotic-resistant pathogens
There are several mechanisms through which bacteria may develop resistance to antibiotics. Among the most important ones are genetic mutations in chromosomal genes and the acquisition of resistance genes through horizontal genetic transfer. Recent research suggests an alternative genetic path: the transfer of large segments of chromosomal DNA between bacteria, thus creating strains with hybrid chromosomes that through one single step may become highly resistant to antibiotics or more virulent.
Two examples of clinical strains that fit this description are ST1193 (Escherichia coli) and ST258 (Klebsiella pneumoniae). Both are multidrug-resistant, highly virulent, and globally widespread. Each strain is a genetic hybrid with at least 20% (>1 Mb) of its chromosome originating in a separate strain. ST1193 has also an interesting genetic architecture: its hybrid chromosome contains 11 non-contiguous fragments of foreign DNA which are thought to have integrated through independent homologous recombination events.
Currently there is no information on the mechanisms of hybrid formation, on the prevalence of hybrid strains among clinical and natural isolates, or on their general importance for antibiotic resistance.
When a conjugative plasmid integrates into the chromosome, it generates an Hfr strain, bacteria with the potential to conjugate chromosomal DNA into a recipient, not only within species but also across genus barriers. The hypothesis of this project is that the phenomenon also occurs in clinical strains with plasmids carrying antibiotic-resistant genes, giving rise to novel hybrids. The frequency of such events is currently unknown. The hypothesis of this project is that the formation of pathogenic strains through processes that create chromosomal hybrids is almost certainly a hugely underappreciated source of genetic variants with novel phenotypic features including resistance to antibiotics.
The major aims of this project are:
- To establish experimental systems to study mechanisms and rates of chromosomal hybrid formation using both laboratory strains (controlled genetics) and clinical isolates (healthcare relevance).
- To investigate the prevalence of chromosomal hybrids among clinical isolates of important bacterial pathogens, and correlations with antibiotic-resistance, including using in silico analysis with bioinformatic and AI pattern recognition software.
- To experimentally evaluate environmental and genetic factors that could enhance or constrain the creation of chromosomal hybrid strains.
Experimentally generated data on hybrids will provide a framework to inform expectations when doing in silicoanalysis of clinical isolates, and will also provide valuable information on the factors that influence rate of formation. In summary, the project will investigate a potentially important cause of genome-wide allelic exchange in bacteria that has the potential to create organisms with novel combinations of selectable phenotypes affecting antibiotic resistance and virulence.
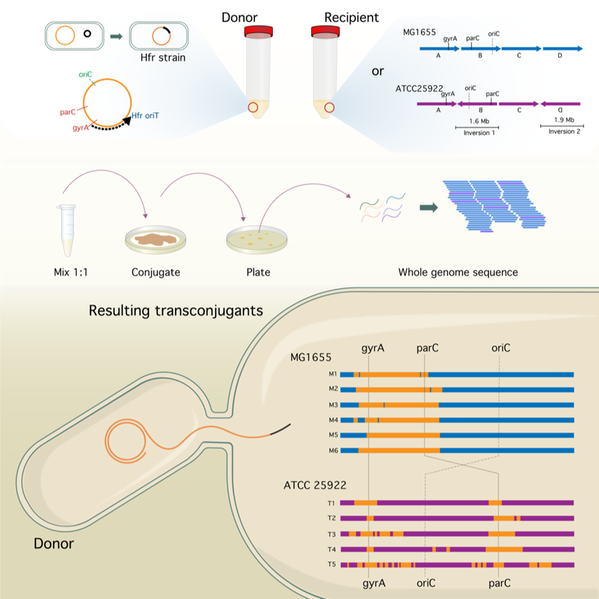
Graphical abstract of one of the ongoing projects
Related published research
- Bartke, K., L. Garoff, D. L. Huseby, G. Brandis and D. Hughes (2020). "Genetic architecture and fitness of bacterial interspecies hybrids." Mol Biol Evol.
- Cavalli, L. L., J. Lederberg and E. M. Lederberg (1953). "An infective factor controlling sex compatibility in Bacterium coli." J Gen Microbiol 8(1): 89-103.
- Chen, L., B. Mathema, J. D. Pitout, F. R. DeLeo and B. N. Kreiswirth (2014). "Epidemic Klebsiella pneumoniae ST258 is a hybrid strain." mBio 5(3): e01355-01314.
- Hayes, W. (1953). "The mechanism of genetic recombination in Escherichia coli." Cold Spring Harb Symp Quant Biol 18: 75-93.
- Matthey, N., S. Stutzmann, C. Stoudmann, N. Guex, C. Iseli and M. Blokesch (2019). "Neighbor predation linked to natural competence fosters the transfer of large genomic regions in Vibrio cholerae." Elife 8.
- Rayssiguier, C., D. S. Thaler and M. Radman (1989). "The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants." Nature 342(6248): 396-401.
- Tchesnokova, V., M. Radey, S. Chattopadhyay, L. Larson, J. L. Weaver, D. Kisiela and E. V. Sokurenko (2019). "Pandemic fluoroquinolone resistant Escherichia coli clone ST1193 emerged via simultaneous homologous recombinations in 11 gene loci." Proc Natl Acad Sci U S A 116(29): 14740-14748.
- Wanner, Barry L. (1986). “Novel Regulatory Mutants of the Phosphate Regulon in Escherichia coli K-12.” J. Mol. Biol. 191, 39-58.
