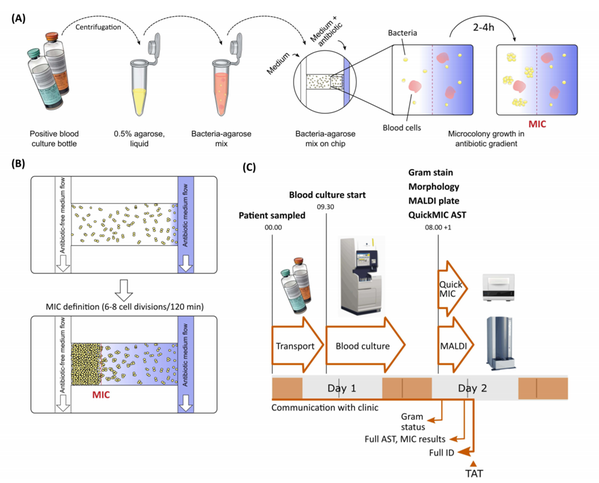
Antibiotic Susceptibility Testing with microfluidics technology
Bacteria are normally rapidly cleared from the blood by the immune system. When the body for some reason cannot remove the cells, bacterial infection may arise, in turn possibly leading to sepsis and septic shock. Mortality in sepsis is very high, but can be reduced by early and appropriate antibiotic treatment. With increasing antimicrobial resistance however, the likelihood of successful treatment is reduced. Traditional antimicrobial susceptibility testing is however slow and inadequate in time-critical disease such as sepsis. There is an urgent need for new, more rapid and accurate antibiotic susceptibility tests. Christer Malmberg's PhD project "Refinement of a robust and rapid microfluidics-based antibiotic susceptibility test with single cell resolution - from prototype to clinical implementation" aims to develop exactly such a test in cooperation with the Uppsala-based biotech company Gradientech AB.
Optical detection system for accurate solid-phase cytometry of very wide-field cell matrices with microfluidic substance gradients
One challenge to improving the speed of phenotypic susceptibility testing is the need to rapidly, accurately and non-invasively capture accurate data from a very large collection of cells, since antibiotic susceptibility is a population-scale measurement. We are investigating a low-magnification, wide-field optical setup with high cell-mass resolution for simultaneously quantifying bacterial growth rates of tens of thousands of bacterial cells growing in antibiotic gradients.
Large-scale validation of a novel rapid microfluidics-based antibiotic susceptibility test - preclinical and clinical verification
Development of new diagnostic methods for diagnosing patients in critical condition, such as sepsis, leads to high demands on reliability and accuracy of the method. Continuous testing with challenging resistance phenotypes is key for optimizing performance. In this project we iteratively test rapid AST test system prototypes to collect pre-clinical and clinical performance data.
Using T2Dx and rapid AST with blood culture presampling for combined ID and AST before blood culture positivity
For improved clinical care all steps of the sepsis diagnostics pipeline must be improved - new culture-free sepsis diagnostic methods directly using patient blood are becoming available, such as the T2Dx system. In this project we investigate linking early sepsis detection and bacterial identification using T2Dx with rapid AST using our in-house system, with the goal to reduce total turn-around times to hours instead of days.
Rapid detection of bacterial co-infections among hospitalized patients with COVID-19 using T2Dx
The current global pandemic has highlighted knowledge gaps in the role of bacterial superinfections in viral pneumonia. Using rapid, molecular detection methods we aim to detect bacterial and fungal co-infections in hospitalized and intensive care patients.
Related published research
- Wistrand-Yuen, Pikkei, Christer Malmberg, Nikos Fatsis-Kavalopoulos, Moritz Lübke, Thomas Tängdén, and Johan Kreuger. 2020. “A Multiplex Fluidic Chip for Rapid Phenotypic Antibiotic Susceptibility Testing.” MBio 11 (1).
- Ungphakorn, Wanchana, Christer Malmberg, Pernilla Lagerbäck, Otto Cars, Elisabet I. Nielsen, and Thomas Tängdén. 2017. “Evaluation of Automated Time-Lapse Microscopy for Assessment of in Vitro Activity of Antibiotics.” Journal of Microbiological Methods 132 (January): 69–75.
- Christer Malmberg, Pikkei Yuen, Johanna Spaak, Otto Cars, Thomas Tängdén, and Pernilla Lagerbäck. 2016. “A Novel Microfluidic Assay for Rapid Phenotypic Antibiotic Susceptibility Testing of Bacteria Detected in Clinical Blood Cultures.” PLOS ONE 11 (12): e0167356.
- Wistrand-Yuen, Pikkei, Christer Malmberg, Thomas Läräng, Thomas Tängdén, and Johan Kreuger. 2016. “Rapid and High-Throughput Phenotypic AST Directly from Positive Blood Cultures with Very High Precision MIC Values.” In EPoster, ECCMID 2019. Amsterdam.
- Yuen, Pikkei, Christer Malmberg, Pernilla Lagerbäck, Otto Cars, and Thomas Tängdén. 2016. “Improved Detection Algorithms for Label-Free Time-Lapse Cytometry of Bacterial Microcolonies for Rapid Antibiotic Susceptibility Testing.” In EPoster, ECCMID 2016. Amsterdam.
