A DNA-encoded compound library-based approach for the discovery of novel Lep-B inhibitors
Antibiotics constitute the cornerstone of modern medicine, and they are one of the most successful class of drugs ever used. However, their effectiveness against infectious diseases is currently being challenged by the emergence of pan-drug resistant bacterial pathogens. Multidrug and even pan-drug resistance bacterial pathogens are threatening the effectiveness of antibiotic therapy worldwide. Hence, there is an urgent need to discover and develop new antibiotics. In our project, we will seek to identify structurally novel inhibitors of LepB, a promising but currently underexplored antibiotic target, which plays an essential role in the secretory pathways of many Gram-positive and Gram-negative bacterial species.
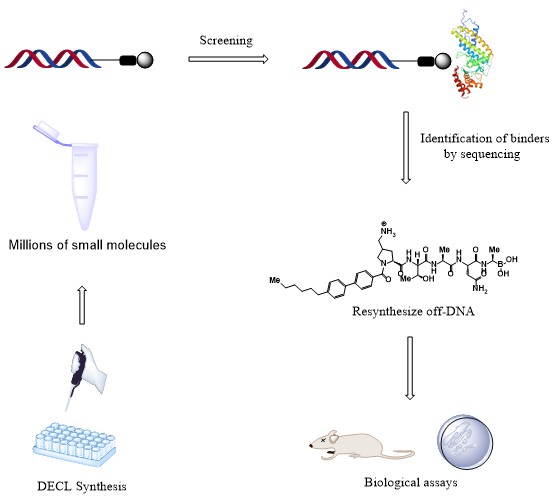
To enable efficient hit discovery, we plan to make use of the DNA-encoded compound library (DECL) technology, which currently constitutes the most efficient and cost-effective method for synthesizing and screening compound libraries. This project is based on previous research carried out at our department, the discovery of an oligopeptide that was found to be a potent inhibitor of E.coli LepB with a promising antibacterial activity against several bacterial strains. However, to improve the toxicity profile of this peptide, we will take advantage of DECL technology in seeking new and safer inhibitors of LepB. We will use the DECL technology to identify new binders of LepB that have been designed based on this oligopeptide. The plan is to develop an extensive collection of potent analogs of this oligopeptide and examine whether we can discover compounds having the desired antibacterial activity with an improved toxicity profile compared to the original oligopeptide. In parallel, we will also do selections from two unbiased peptide-based DECLs with the aim of identifying novel series of LepB binders.
Related published research
- Dalbey, Ross E., Mark O. Lively, Sierd Bron, and Jan Maarten Van Dijl. “The Chemistry and Enzymology of the Type I Signal Peptidases.” Protein Science 6, no. 6 (1997): 1129–38.
- Mannocci, Luca, Yixin Zhang, Jörg Scheuermann, Markus Leimbacher, Gianluca De Bellis, Ermanno Rizzi, Christoph Dumelin, Samu Melkko, and Dario Neri. “High-Throughput Sequencing Allows the Identification of Binding Molecules Isolated from DNA-Encoded Chemical Libraries.” Proceedings of the National Academy of Sciences105, no. 46 (November 18, 2008): 17670–75.
- Oschmann, Michael, Linus Johansson Holm, Monireh Pourghasemi‐Lati, and Oscar Verho. “Synthesis of Elaborate Benzofuran‐2‐carboxamide Derivatives through a Combination of 8-Aminoquinoline Directed C–H Arylation and Transamidation Chemistry.” Molecules 25, no. 2 (2020): 1–14.
- Rao C.V., Smitha, Evelien De Waelheyns, Anastassios Economou, and Jozef Anné. “Antibiotic Targeting of the Bacterial Secretory Pathway.” Biochimica et Biophysica Acta - Molecular Cell Research 1843, no. 8 (2014): 1762–83.
- Rosa, Maria De, Lu Lu, Edouard Zamaratski, Natalia Szałaj, Sha Cao, Henrik Wadensten, Lena Lenhammar, et al. “Design, Synthesis and in Vitro Biological Evaluation of Oligopeptides Targeting E. Coli Type I Signal Peptidase (LepB).” Bioorganic and Medicinal Chemistry 25, no. 3 (2017): 897–911.
- Salamon, Hazem, Mateja Klika Škopić, Kathrin Jung, Olivia Bugain, and Andreas Brunschweiger. “Chemical Biology Probes from Advanced DNA-Encoded Libraries.” ACS Chemical Biology 11, no. 2 (2016): 296–307.
- Szałaj, Natalia, Lu Lu, Andrea Benediktsdottir, Edouard Zamaratski, Sha Cao, Gustav Olanders, Charles Hedgecock, et al. “Boronic Ester-Linked Macrocyclic Lipopeptides as Serine Protease Inhibitors Targeting Escherichia Coli Type I Signal Peptidase.” European Journal of Medicinal Chemistry 157 (2018): 1346–60.
