Research projects in Ivarsson Group
In a cell, there are more than 100,000 protein-protein interactions taking place at any given moment, and these are crucial for the cell's function. When errors occur in protein interactions, for example mutation of a binding surface, this can lead to diseases such as various types of cancer. Today, we only know a fraction of all interactions, and great efforts are being made to map them.
Our group has several on-going projects with focus on molecular interactions. We use proteomic peptide phage display to identify interactions of potential biological relevance, bioinformatics to filter for relevant binders and in vitro affinity measurements and cell-based assays for detailed analysis of key interactions. When appropriate, we also probe the interplay between peptide and phospholipid binding. With our research, we contribute new knowledge about the interactions that can take place in a human cell in a healthy and diseased state.
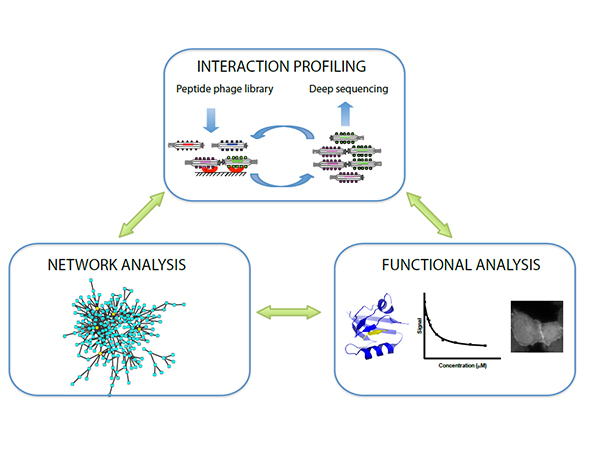
Interaction profiling through proteomic peptide phage display
A significant part of the human proteome is intrinsically disordered. These regions are enriched in short motifs serving as docking sites for peptide binding modules. Peptide-motifs interactions are crucial for the wiring of signaling pathways. These important but transient interactions are difficult to capture through most conventional high-throughput methods. We apply a novel approach for the large-scale profiling of domain-motifs interactions called Proteomic Peptide Phage Display (ProP-PD) (Ivarsson et al, 2014, Sundell & Ivarsson, 2014). ProP-PD can be used to search for protein-motif interactions of potential biological relevance on a proteome wide scale and it provides information that is complementary to other high-throughput methods.
