Metabolomics

Research with a focus on ensuring high data quality and developing new and improved methods and to improve mechanistic understanding, prognostics and diagnostics for various forms of cancer.
Metabolomics is the simultaneous analysis of all small molecular molecules, metabolites, in a defined biological system where no specific substance or group is of particular interest in advance. Thus, Metabolomics can be described as generating hypotheses, which are consequently tested in follow-up studies, for example through targeted, bioanalytical analyses.

Our research in metabolomics focuses on ensuring high data quality and developing new and improved methods. We also collaborate with groups in projects that, among other things, aim to improve mechanistic understanding, prognostics and diagnostics for various forms of cancer.
Data quality in untargeted metabolomics
While the field of bioanalytical chemistry is well developed in terms of quality assurance, the idea of performing similar validation in untargeted metabolomics studies is staggering due to the huge number of substances that is being measured in each study. Consequently, the assessment of data quality and validation standardization is under-developed. At the same time data of the highest possible quality is crucial for arriving at correct hypotheses, which is why we dedicate much effort to the development of suitable validation for untargeted metabolomics methods.
Read our review, LC–MS based global metabolite profiling: the necessity of high data quality (Metabolomics (2016) 12:114), for an extended discussion on the topic as well as information on current best practices and suggestions.
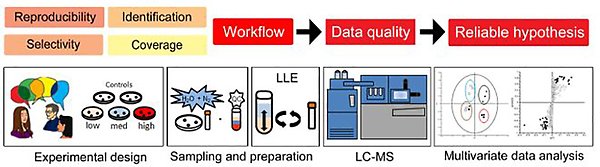
Analytical methods for metabolomics
Untargeted metabolomics imply the simultaneous measurement of hundreds to thousands of metabolites. While this might not entail all metabolites in a given sample, it places heavy demands on all stages of the analysis in terms of foremost reproducibility and selectivity.
These demands need to affect the entire study, from experimental design, through sampling, sample preparation, separation, detection, data processing and statistical analysis to biological hypothesis generation.We therefore strive at developing methods better suited to the needs of each study; not only regarding reporducibility and selectivity, but also to meet demands on e.g. sample preparation, separation or detection.
For example, cell quenching and harvesting using nitrogen snap freezing as opposed to methanol washing allows for more tailored sample preparation as described in Metabolic profiling of epithelial ovarian cancer cell lines: evaluation of harvesting protocols for profiling using NMR spectroscopy (Bioanalysis (2015) 7:157).
Applied studies
We collaborate with several research groups within Uppsala University, at Uppsala Academic Hospital, Karolinska Institutet and Queen Mary University of London. Many studies are focused around the mechanistics, prognostics and diagnostics of various forms of cancer; other studies concern neurotoxins or nutrition interventions.
Samples
We routinely analyze biofluids like blood or urine samples, as well as harvested in vitro cell samples, although other samples are handled as well.
Facilities
We use both NMR, UPLC-HRMS and UPLC-MS/MS depending on the needs of each respective study. To that end, we currently employ:
- Bruker Avance 600 MHz with cryoprobe
- Waters Acquity I-class UPLC
- Waters Synapt G2-S Q-TOF
- Waters QuattroMicro Tandem Quadrupole
In the press
- New study to provide more effective treatment for primary liver cancer (Uppsala University)
Kontakt:
Jenny Nilsson, PhD Student
Department of Medicinal Chemistry
Jenny.M.Nilsson@uu.se
Project members
Contact
- Mikael Hedeland
- Institutionen för läkemedelskemi, 070-657 1663
