Dan I Andersson research group
Tempo and mode of bacterial evolution
Our research addresses the mechanisms and dynamics of evolution in bacteria and how various factors such as the extent and type of genetic variation, strength of selection pressures, compensatory mutations and population dynamics affect the tempo and mode of adaptive evolution. Our research is focused on three different areas:
A) analysis of the various genetic factors that affect genome stability and variability in bacteria; B) analysis of the factors that influence the dynamics of the evolution of antibiotic resistance development; C) analysis of antibiotic combinations treatments and the interplay between antibiotics. We study these problems in several bacterial species using a combination of methods, including experimental evolution, bacterial genetics, molecular biology, biochemistry, next generation sequencing and mathematical modeling.
A. Genome variability and stability.

Figure 1. Genome varability via horizontal gene transfer, de novo gene birth and duplication-divergence mechanisms.
The long-term goal of this project is to examine the major factors that influence the tempo and mode of bacterial evolution. In particular we are interested in the evolutionary and mechanistic factors that influence genome stability and variability. We use several microorganisms such as E. coli and S. Typhimurium as model systems to:
- Examine how the extent and type of genetic variation affects bacterial fitness and rates of adaptive evolution.
- Examine the role of gene amplification in adaptive responses (e.g. during exposure to antimicrobial drugs) and in the evolution of novel genes.
- Examine the fitness effects and constraints on horizontal gene transfer.
- Examine the distribution of fitness effects of random mutations and its impact on adaptive evolution.
- Examine the impact of genetic diversity on rates of adaptive evolution.
- Examine the distribution of gene copy number variation withing populations and the impact on daptive evolution.
B. Mechanisms and dynamics of the evolution of antibiotic resistance.
%20dan%20bild%202.jpg)
Figure 2. Sub-inhibitory levels of antibiotics leads to resistance selection and de novo resistance development.
The overall objective of this project is to understand how antibiotic resistance affects the fitness, virulence and transmission of various pathogenic bacteria (e.g. S. typhimurium, E. coli, S. aureus, M. tuberculosis, K. pneumoniae and P. aeruginosa) and which factors determine how rapidly resistance develops in bacterial populations. Our main aims are to:
- Determine how various types of resistance mechanisms affect bacterial fitness and virulence.
- Determine how bacteria can compensate for resistance-conferred fitness costs.
- Examine the importance of genetic epistasis on the rate and trajectory of multi-drug-resistance development and compensatory evolution.
- Determine in animal models and human volunteers how antibiotic resistance affects bacterial transmission.
- Examine the feasibility of reversion of resistance by determining if reduced antibiotic use in community settings may result in a reduced frequency of resistance.
- Examine how ultra-low levels of antibiotics affect enrichment and de novo selection for resistant mutants in various types of aquatic environments.
- Examine mechanisms of gene copy number variation and their impact on antibiotic resistance development
- Assess the prevalence, mechanisms and clinical relevance of antibiotic heteroresistance
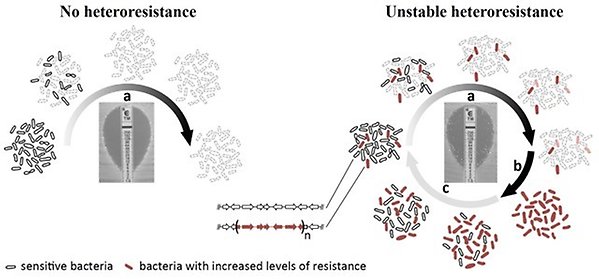
Figure 3. Heteroresistance is a bacterial phenotype where, within a susceptible main population, there exist small resistant subpopulation(s). In a non-heteroresistant isolate all bacteria will die at a certain antibiotic concentration but in a heteroresistant isolate a subpopulation will survive and grow at a substantially higher concentration of antibiotic. Unstable heteroresistance involves the amplification of antibiotic resistance genes leading to an increased gene dosage and increased antibiotic resistance.
C. Antibiotic combination treatments and detection of interplay between antibiotics.
Resistance is increasingly challenging the effectiveness of antibiotic treatments, making the use of antibiotic combinations a common strategy in clinics to combat this problem. However, not all combinations work effectively—some can interfere with each other’s activity, while others can enhance it. Understanding the specific interactions between antibiotics, whether they hinder or help each other, can lead to more effective therapies and better patient outcomes.
Our research is aimed at understanding these interactions, using innovative diagnostic techniques to uncover both the clinical and mechanistic aspects of how antibiotics interact. By analysing how different antibiotics work together, we can identify synergies where the combined effect is greater than the sum of their individual actions, and antagonisms where the combination is less effective. Beyond identifying these interactions, our study also explores the fundamental processes that underpin these effects, providing deeper insights into the molecular and cellular mechanisms that drive antibiotic interactions.

Figure 4. Antagonism and synergy on the same plate (C synergizes with A and B while A and B antagonize). In the darker areas bacteria are growing whereas in the lighter areas bacterial growth is inhibited.
Publications
Part of Nature Ecology & Evolution, p. 1321-1330, 2018
A broad spectrum anti-bacterial peptide with an adjunct potential for tuberculosis chemotherapy
Part of Scientific Reports, 2021
- DOI for A broad spectrum anti-bacterial peptide with an adjunct potential for tuberculosis chemotherapy
- Download full text (pdf) of A broad spectrum anti-bacterial peptide with an adjunct potential for tuberculosis chemotherapy
Part of Journal of Clinical Microbiology, p. 425-432, 2015
- DOI for A General Method for Rapid Determination of Antibiotic Susceptibility and Species in Bacterial Infections
- Download full text (pdf) of A General Method for Rapid Determination of Antibiotic Susceptibility and Species in Bacterial Infections
Part of Journal of Antimicrobial Chemotherapy, p. 3051-3060, 2015
Part of Frontiers in Cellular and Infection Microbiology, 2022
- DOI for A Microfluidic Chip for Studies of the Dynamics of Antibiotic Resistance Selection in Bacterial Biofilms
- Download full text (pdf) of A Microfluidic Chip for Studies of the Dynamics of Antibiotic Resistance Selection in Bacterial Biofilms

