Linus Sandegren research group

Dynamics of plasmid-borne antibiotic resistance
If you want to work with us, contact Linus Sandegren for available positions.
We study different aspects of bacterial evolution concerning antibiotic resistance development and bacterial virulence to understand better what factors and molecular mechanisms affect bacterial development of resistance and infection. We are situated in a very dynamic research environment with several research groups that work with complementary research questions and methodologies, and we collaborate with researchers at several other universities and hospitals.
Dynamics of plasmid-borne antibiotic resistance
We study fundamental aspects of how resistance plasmids are maintained and disseminated between pathogenic bacteria and how they serve as platforms for the evolution of antibiotic resistance. The main focus is to understand how factors such as stability, mobility, positive selection, and fitness cost influence the evolutionary success of plasmids. The experimental systems we use are based on clinical multi-resistance plasmids encoding extended-spectrum β-lactamases (ESBLs) in enteric bacteria (Escherichia coli and Klebsiella pneumoniae) that pose an increasing clinical problem by providing bacteria with resistance to the most used antibiotics today, β-lactams and β-lactamase inhibitors.
Five main themes are of particular interest in these studies:
- What impact do low levels of antibiotics have on the spread, selection, and maintenance of multi-resistance plasmids?
- What plasmid factors cause a fitness cost on the host cell, and how can the bacterium alleviate the fitness cost of plasmid carriage?
- How common are gene amplifications, how do they help bacteria survive antibiotics, and does the dynamics of gene amplification on plasmids accelerate the evolution of new resistance?
- How does bacterial growth influence resistance development and plasmid dynamics in biofilms compared to planktonic growth?
From these studies, we learn how bacterial cells and plasmids co-evolve and how the selection of new resistance can accelerate through gene amplification at different antibiotic concentrations. Such knowledge can be used to design antibiotic treatment regimens that limit resistance selection and minimize the potential for new resistance to evolve.
Combinatorial effects of plasmid-borne β-lactamases on resistance development to different combination therapies with β-lactamase inhibitors and β-lactam antibiotics
The primary resistance mechanism against β-lactams among Gram-negative bacteria is the production of β-lactamases, enzymes that degrade β-lactams. By combining the β-lactam antibiotics with a β-lactamase inhibitor, which inactivates the β-lactamase, successful treatment of infections can be regained. Unfortunately, bacteria can circumvent the effect of the combination therapy by overexpressing existing β-lactamases or by altered porin expression, reducing antibiotic entry into the bacterium. We study how bacterial strains carrying resistance plasmids with multiple β-lactamase genes affect the combination therapy and evolution of novel resistance to β-lactam/ β-lactamase inhibitor combinations. The role of gene amplification and evolution of the catalytic properties of β-lactamases is of particular focus.
Within-host evolution of Klebsiella pneumoniae - How does an opportunistic pathogen specialize?
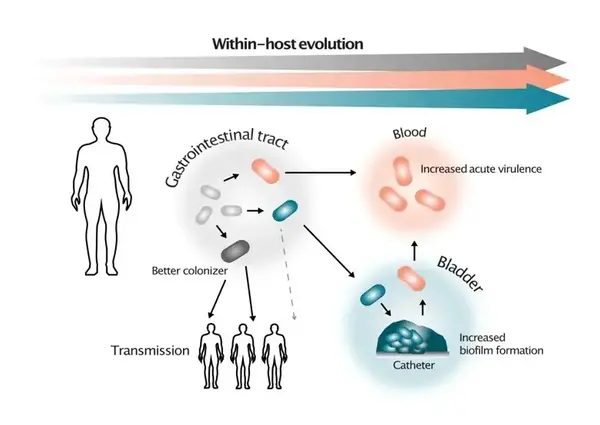
Evolutionary scenarios for in-host selection of subpopulations with altered virulens. Illustrations by Greta Zaborskyte.
How bacteria balance between benign colonization of the human host and virulence is a central question in infection biology. Despite long-standing advances in the molecular understanding of virulence factors in specific pathogens, there are still significant gaps in our knowledge of the selective forces and spontaneous genetic changes that lead to short-term bacterial adaptation to different niches in their host. A key question is the balance between optimal fitness in the colonizing niche and the potential to invade and use new sites in the body for proliferation. In this project, we combine genotypic and phenotypic characterization of a unique collection of Klebsiella pneumoniae clonal bacterial isolates from a large hospital outbreak and isolates of global high-risk clones. Through experimental evolution, advanced bacterial genetics, and novel biofilm- and virulence models, we determine critical steps and selective forces of bacterial within-host evolution of this important nosocomial pathogen. K. pneumoniae is classically viewed as an opportunistic pathogen. However, lately, hypervirulent clones have spread globally, and it has been proposed that the success of these high-risk clones comes from the parallel evolution of virulence/colonization and antibiotic resistance.
Klebsiella pneumoniae biofilm evolution and plasmid dynamics
Bacteria can grow as communities attached to surfaces – biofilms. Biofilm growth is inherent in opportunistic infections, where the protected bacterial community can withstand antibiotic treatments and immune attacks. Indwelling medical devices that support biofilm growth often facilitate the transition between colonization and infection. For classical K. pneumoniae, this translates into hospital-acquired urinary tract infections or ventilator-associated pneumonia exacerbated by the presence of urinary catheters or endotracheal tubes. In addition, bacterial plasmids and biofilms seem to be in a complex relationship. For instance, plasmids can provide structural components for biofilms (e.g., fimbriae, extracellular DNA), and biofilms can, in turn, affect plasmid biology by affecting conjugation frequency or copy number heterogeneity in the more complex and compartmentalized structure of the biofilm.
This project addresses how K. pneumoniae can increase biofilm capacity by experimentally evolving surface-attached biofilms to mimic catheter-associated infections. We are also using Klebsiella pneumoniae in vitro biofilms as a model for studying plasmid biology in surface-attached biofilms. The overall goals are to find out how antibiotic treatment, especially at low concentrations, affects bacteria in biofilms and the plasmid-biofilm relationship, focusing on the evolution of plasmid-borne antibiotic resistance mechanisms.
We have developed an in vitro biofilm model system compatible with live in situ imaging to monitor the invasion and spread of resistance plasmids in the biofilms exposed to antibiotics. The knowledge gained in this project will give insights into more efficient treatment of biofilm-related infections.
CAD files for the FlexiPeg biofilm system can be found here.
